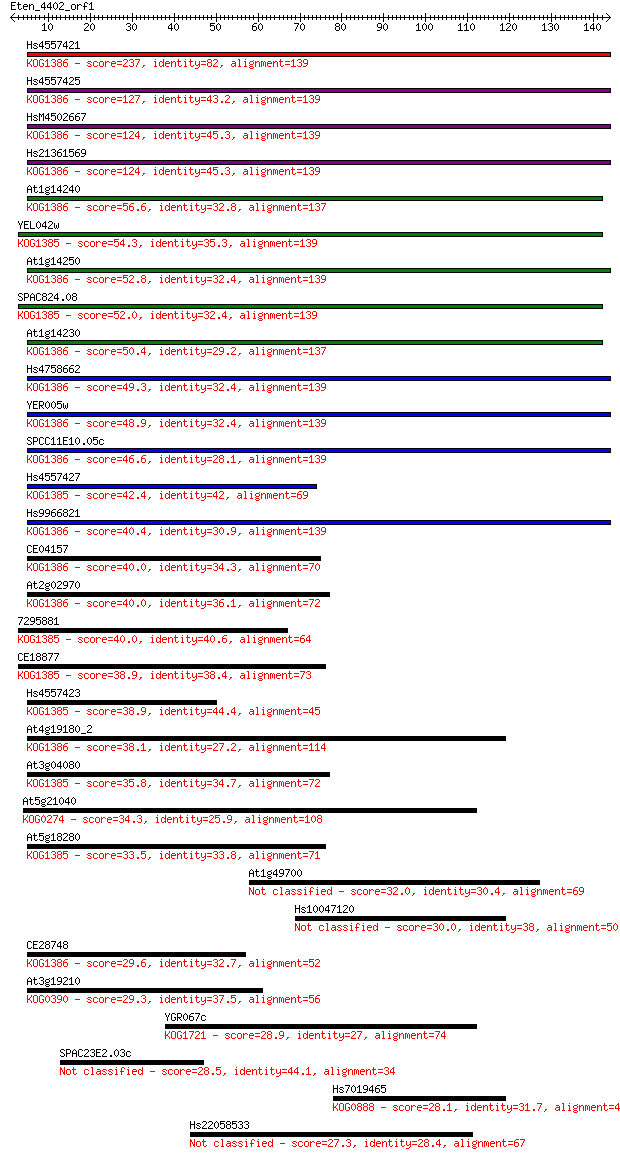
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4402_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
Hs4557421 237 5e-63
Hs4557425 127 7e-30
HsM4502667 124 6e-29
Hs21361569 124 6e-29
At1g14240 56.6 1e-08
YEL042w 54.3 8e-08
At1g14250 52.8 2e-07
SPAC824.08 52.0 3e-07
At1g14230 50.4 1e-06
Hs4758662 49.3 2e-06
YER005w 48.9 3e-06
SPCC11E10.05c 46.6 2e-05
Hs4557427 42.4 3e-04
Hs9966821 40.4 0.001
CE04157 40.0 0.001
At2g02970 40.0 0.001
7295881 40.0 0.002
CE18877 38.9 0.003
Hs4557423 38.9 0.004
At4g19180_2 38.1 0.006
At3g04080 35.8 0.027
At5g21040 34.3 0.074
At5g18280 33.5 0.12
At1g49700 32.0 0.42
Hs10047120 30.0 1.6
CE28748 29.6 1.9
At3g19210 29.3 2.4
YGR067c 28.9 3.1
SPAC23E2.03c 28.5 4.2
Hs7019465 28.1 5.1
Hs22058533 27.3 9.9
> Hs4557421
Length=472
Score = 237 bits (604), Expect = 5e-63, Method: Compositional matrix adjust.
Identities = 114/139 (82%), Positives = 122/139 (87%), Gaps = 0/139 (0%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLARALQT 64
+DLGGASTQITFETTSP ED A+EVQL LYGQ YRVYTHSFLCYGRDQVL+RLLA ALQT
Sbjct 200 MDLGGASTQITFETTSPAEDRASEVQLHLYGQHYRVYTHSFLCYGRDQVLQRLLASALQT 259
Query 65 HGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCRSLVSQLFN 124
HG HPCWPRG+ST V+L DVY+SPCT AQRPQ FN S RVSL+GSSDP LCR LVS LF+
Sbjct 260 HGFHPCWPRGFSTQVLLGDVYQSPCTMAQRPQNFNSSARVSLSGSSDPHLCRDLVSGLFS 319
Query 125 SSSCHFSRCSFNGVFQPPV 143
SSC FSRCSFNGVFQPPV
Sbjct 320 FSSCPFSRCSFNGVFQPPV 338
> Hs4557425
Length=529
Score = 127 bits (319), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 60/146 (41%), Positives = 94/146 (64%), Gaps = 7/146 (4%)
Query 5 LDLGGASTQITFETTSPTE-DPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLARALQ 63
LDLGGASTQI+F + + ++ +Q+ LYG Y +YTHSF CYGR++ ++ LA LQ
Sbjct 218 LDLGGASTQISFVAGEKMDLNTSDIMQVSLYGYVYTLYTHSFQCYGRNEAEKKFLAMLLQ 277
Query 64 T-----HGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCRSL 118
H ++PC+PR YS + V++S CT QRP+++N + ++ G+ DP+LC+
Sbjct 278 NSPTKNHLTNPCYPRDYSISFTMGHVFDSLCTVDQRPESYNPNDVITFEGTGDPSLCKEK 337
Query 119 VSQLFNSSSCHFSR-CSFNGVFQPPV 143
V+ +F+ +CH CSF+GV+QP +
Sbjct 338 VASIFDFKACHDQETCSFDGVYQPKI 363
> HsM4502667
Length=510
Score = 124 bits (311), Expect = 6e-29, Method: Composition-based stats.
Identities = 63/144 (43%), Positives = 90/144 (62%), Gaps = 7/144 (4%)
Query 5 LDLGGASTQITFETTSPT-EDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLARALQ 63
LDLGGASTQ+TF + T E P N +Q RLYG+ Y VYTHSFLCYG+DQ L + LA+ +Q
Sbjct 212 LDLGGASTQVTFVPQNQTIESPDNALQFRLYGKDYNVYTHSFLCYGKDQALWQKLAKDIQ 271
Query 64 THGSH----PCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCRSLV 119
+ PC+ GY V + D+Y++PCT +R + + + G + C +
Sbjct 272 VASNEILRDPCFHPGYKKVVNVSDLYKTPCT--KRFEMTLPFQQFEIQGIGNYQQCHQSI 329
Query 120 SQLFNSSSCHFSRCSFNGVFQPPV 143
+LFN+S C +S+C+FNG+F PP+
Sbjct 330 LELFNTSYCPYSQCAFNGIFLPPL 353
> Hs21361569
Length=517
Score = 124 bits (311), Expect = 6e-29, Method: Composition-based stats.
Identities = 63/144 (43%), Positives = 90/144 (62%), Gaps = 7/144 (4%)
Query 5 LDLGGASTQITFETTSPT-EDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLARALQ 63
LDLGGASTQ+TF + T E P N +Q RLYG+ Y VYTHSFLCYG+DQ L + LA+ +Q
Sbjct 219 LDLGGASTQVTFVPQNQTIESPDNALQFRLYGKDYNVYTHSFLCYGKDQALWQKLAKDIQ 278
Query 64 THGSH----PCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCRSLV 119
+ PC+ GY V + D+Y++PCT +R + + + G + C +
Sbjct 279 VASNEILRDPCFHPGYKKVVNVSDLYKTPCT--KRFEMTLPFQQFEIQGIGNYQQCHQSI 336
Query 120 SQLFNSSSCHFSRCSFNGVFQPPV 143
+LFN+S C +S+C+FNG+F PP+
Sbjct 337 LELFNTSYCPYSQCAFNGIFLPPL 360
> At1g14240
Length=483
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 45/151 (29%), Positives = 66/151 (43%), Gaps = 27/151 (17%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQLRLYGQ-RYRVYTHSFLCYGRDQVLRRLLARALQ 63
++LGGAS Q+TF S P + YG Y +Y+HSFL YG+D L++LL +
Sbjct 221 VELGGASAQVTF--VSSEHVPPEYSRTIAYGNISYTIYSHSFLDYGKDAALKKLLEKLQN 278
Query 64 THGS-------HPCWPRGYSTHVMLWDV----YESPCTAAQRPQAFNRSTRVSLAGSSDP 112
+ S PC P+GY ++D Y S A + + SL + +
Sbjct 279 SANSTVDGVVEDPCTPKGY-----IYDTNSKNYSSGFLADE------SKLKGSLQAAGNF 327
Query 113 ALCRSLVSQLFNS--SSCHFSRCSFNGVFQP 141
+ CRS L +C + CS F P
Sbjct 328 SKCRSATFALLKEGKENCLYEHCSIGSTFTP 358
> YEL042w
Length=518
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 49/162 (30%), Positives = 66/162 (40%), Gaps = 35/162 (21%)
Query 3 APLDLGGASTQITFETTSPTE----DPANEVQLRLYGQRYRVYTHSFLCY----GRDQVL 54
A DLGG STQI FE T P D ++ L+ + Y +Y S L Y GR++V
Sbjct 242 AVFDLGGGSTQIVFEPTFPINEKMVDGEHKFDLKFGDENYTLYQFSHLGYGLKEGRNKVN 301
Query 55 RRLLARAL-----------QTHG-SHPCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRST 102
L+ AL +TH S PC P + T + +
Sbjct 302 SVLVENALKDGKILKGDNTKTHQLSSPCLPPKVNA------------TNEKVTLESKETY 349
Query 103 RVSLAGSSDP--ALCRSLVSQLFNS-SSCHFSRCSFNGVFQP 141
+ G +P A CR L ++ N + C CSFNGV QP
Sbjct 350 TIDFIGPDEPSGAQCRFLTDEILNKDAQCQSPPCSFNGVHQP 391
> At1g14250
Length=488
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 68/154 (44%), Gaps = 26/154 (16%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQLRL-YGQ-RYRVYTHSFLCYGRDQVLRRLLARAL 62
++LGGAS Q+TF P+E E + YG Y +Y+HSFL +G+D +LL +L
Sbjct 222 VELGGASAQVTF---VPSEHVPPEFSRTISYGNVSYTIYSHSFLDFGQDAAEDKLL-ESL 277
Query 63 QTHGS---------HPCWPRGYSTHVMLWDVYESPCTAA--QRPQAFNRSTRVSLAGSSD 111
Q + PC P+GY ++D + ++ F S +V AG D
Sbjct 278 QNSVAASTGDGIVEDPCTPKGY-----IYDTHSQKDSSGFLSEESKFKASLQVQAAG--D 330
Query 112 PALCRSLVSQLFNS--SSCHFSRCSFNGVFQPPV 143
CRS + +C + CS F P +
Sbjct 331 FTKCRSATLAMLQEGKENCAYKHCSIGSTFTPNI 364
> SPAC824.08
Length=556
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 45/155 (29%), Positives = 71/155 (45%), Gaps = 23/155 (14%)
Query 3 APLDLGGASTQITFETTSPTE-----DPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRL 57
A +DLGGASTQ+ FE ++ D ++ L G++Y +Y HS L YG + R+L
Sbjct 281 AVMDLGGASTQLVFEPRFASDGESLVDGDHKYVLDYNGEQYELYQHSHLGYGLKEA-RKL 339
Query 58 LARALQTHGS---HPCWPRGYSTHVMLWDVYESPC----TAAQRPQAFNRSTRVSLAGSS 110
+ + + + G ST ++ PC + P + + ++ V G S
Sbjct 340 IHKFVLNNAEALKESLELLGDSTSII------HPCLHLNASLTHPDSKSEASEVVFVGPS 393
Query 111 DPAL---CRSLVSQ-LFNSSSCHFSRCSFNGVFQP 141
L CR + + L+ +C CSFNGV QP
Sbjct 394 LAHLSLQCRGIAEKALYKDKNCPVRPCSFNGVHQP 428
> At1g14230
Length=519
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 40/149 (26%), Positives = 66/149 (44%), Gaps = 22/149 (14%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQLRLYGQ-RYRVYTHSFLCYGRDQVLRRLLARALQ 63
++LGGAS Q+TF +T P+ + YG Y +Y+HSFL +G+D + L+ +L
Sbjct 217 VELGGASAQVTFVSTELV--PSEFSRTLAYGNVSYNLYSHSFLDFGQDAAQEK-LSESLY 273
Query 64 THGSH---------PCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPAL 114
++ PC P+GY L + P A + + +L + + +
Sbjct 274 NSAANSTGEGIVPDPCIPKGYILETNLQK--DLPGFLADKGK-----FTATLQAAGNFSE 326
Query 115 CRSLVSQLFNSS--SCHFSRCSFNGVFQP 141
CRS + C + RCS +F P
Sbjct 327 CRSAAFAMLQEEKGKCTYKRCSIGSIFTP 355
> Hs4758662
Length=616
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/166 (27%), Positives = 66/166 (39%), Gaps = 41/166 (24%)
Query 5 LDLGGASTQITFE---TTSPTEDPANEVQLRLYGQ------------RYRVYTHSFLCYG 49
LD+GG STQI +E T S EV L + YRVY +FL +G
Sbjct 270 LDMGGVSTQIAYEVPKTVSFASSQQEEVAKNLLAEFNLGCDVHQTEHVYRVYVATFLGFG 329
Query 50 ----RDQVLRRLLARALQTH-------GSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAF 98
R + R+ A +Q + G P P Y PC
Sbjct 330 GNAARQRYEDRIFANTIQKNRLLGKQTGLTPDMP------------YLDPCLPLDIKDEI 377
Query 99 NRSTR-VSLAGSSDPALCRSLVSQLFNSSSCHFSRCSFNGVFQPPV 143
++ + + L G+ D LCR + N ++ ++ S NGV+QPP+
Sbjct 378 QQNGQTIYLRGTGDFDLCRETIQPFMNKTNE--TQTSLNGVYQPPI 421
> YER005w
Length=630
Score = 48.9 bits (115), Expect = 3e-06, Method: Composition-based stats.
Identities = 45/164 (27%), Positives = 65/164 (39%), Gaps = 43/164 (26%)
Query 5 LDLGGASTQITFETTSPTE-----DPANEVQLRLYG---QRYRVYTHSFLCYGRDQVLRR 56
+D+GGASTQI F E D + LR Q++ V+ ++L +G +Q RR
Sbjct 183 MDMGGASTQIAFAPHDSGEIARHRDDIATIFLRSVNGDLQKWDVFVSTWLGFGANQARRR 242
Query 57 LLARALQT----------------HGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAFNR 100
LA+ + T + + PC PRG ST D
Sbjct 243 YLAQLINTLPENTNDYENDDFSTRNLNDPCMPRGSSTDFEFKD----------------- 285
Query 101 STRVSLAGSSDPALC-RSLVSQLFNSSSCHFSRCSFNGVFQPPV 143
T +AGS + C +S+ L + C C FNGV P +
Sbjct 286 -TIFHIAGSGNYEQCTKSIYPLLLKNMPCDDEPCLFNGVHAPRI 328
> SPCC11E10.05c
Length=572
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 39/148 (26%), Positives = 64/148 (43%), Gaps = 25/148 (16%)
Query 5 LDLGGASTQITFETTSPTE-----DPANEVQLRLY-GQR--YRVYTHSFLCYGRDQVLRR 56
LD+GGAS QI FE P++ D + V + L GQ+ Y ++ ++L +G ++ RR
Sbjct 171 LDMGGASVQIAFE-LPPSQLKNYKDSISTVHIGLQNGQQLEYPLFVTTWLGFGANEAYRR 229
Query 57 LLARALQTHGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCR 116
L +++ + + PC+ R + + AG+ D C
Sbjct 230 YLGLLIESENGK------------VGNTLSDPCSLRGRTYDID---GIEFAGTGDLKQCL 274
Query 117 SLVSQLFNSSS-CHFSRCSFNGVFQPPV 143
L L N C C+F+G+ PPV
Sbjct 275 KLTYNLLNKDKPCSMDPCNFDGISIPPV 302
> Hs4557427
Length=428
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 38/79 (48%), Gaps = 11/79 (13%)
Query 5 LDLGGASTQIT----FETTSPTEDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLAR 60
LDLGGASTQIT FE T ++ Y++YTHS+L +G + R
Sbjct 198 LDLGGASTQITFLPQFEKTLEQTPRGYLTSFEMFNSTYKLYTHSYLGFGL-KAARLATLG 256
Query 61 ALQTHGSH------PCWPR 73
AL+T G+ C PR
Sbjct 257 ALETEGTDGHTFRSACLPR 275
> Hs9966821
Length=604
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 43/166 (25%), Positives = 67/166 (40%), Gaps = 41/166 (24%)
Query 5 LDLGGASTQITFETTSPT------EDPANEVQLRLYG---------QRYRVYTHSFLCYG 49
LD+GGAS QI +E + T ++ A ++ L + YRVY +FL +G
Sbjct 257 LDMGGASLQIAYEVPTSTSVLPAKQEEAAKILLAEFNLGCDVQHTEHVYRVYVTTFLGFG 316
Query 50 --------RDQVLRRLLARAL---QTHGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAF 98
D VL L + Q G P P + PC
Sbjct 317 GNFARQRYEDLVLNETLNKNRLLGQKTGLSPDNP------------FLDPCLPVGLTDVV 364
Query 99 NRSTRV-SLAGSSDPALCRSLVSQLFNSSSCHFSRCSFNGVFQPPV 143
R+++V + G D C +++S L S+ S+ S NG++Q P+
Sbjct 365 ERNSQVLHVRGRGDWVSCGAMLSPLLARSNT--SQASLNGIYQSPI 408
> CE04157
Length=485
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 43/79 (54%), Gaps = 9/79 (11%)
Query 5 LDLGGASTQITFETTSPTE--DPANEVQLRLYG------QRYRVYTHSFLCYGRDQVLRR 56
+D+GGAS QI FE + E + N ++ L +Y++Y+ +FL YG ++ L++
Sbjct 180 IDMGGASVQIAFEIANEKESYNGGNVYEINLGSIETNEDYKYKIYSTTFLGYGANEGLKK 239
Query 57 LLARALQTHGSH-PCWPRG 74
+++ S+ C PRG
Sbjct 240 YENSLVKSGNSNDSCSPRG 258
> At2g02970
Length=516
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 41/84 (48%), Gaps = 13/84 (15%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLARAL-Q 63
++LGGAS Q+TF ++ P P + Y +Y+HSFL +G++ +L L +
Sbjct 219 VELGGASAQVTFVSSEPMP-PEFSRTISFGNVTYNLYSHSFLHFGQNAAHDKLWGSLLSR 277
Query 64 THGS-----------HPCWPRGYS 76
H S PC P+GY+
Sbjct 278 DHNSAVEPTREKIFTDPCAPKGYN 301
> 7295881
Length=464
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 26/68 (38%), Positives = 36/68 (52%), Gaps = 10/68 (14%)
Query 3 APLDLGGASTQITFETTSPTEDPANEVQLR---LYGQRYRVYTHSFLCYGRDQVLRRLLA 59
A LDLGG STQ+TF T P + P + + ++ V+THS+L G + A
Sbjct 227 AALDLGGGSTQVTFSPTDPDQVPVYDKYMHEVVTSSKKINVFTHSYLGLG------LMAA 280
Query 60 R-ALQTHG 66
R A+ THG
Sbjct 281 RHAVFTHG 288
> CE18877
Length=479
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 28/84 (33%), Positives = 39/84 (46%), Gaps = 13/84 (15%)
Query 3 APLDLGGASTQITFETTSP---TEDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLA 59
A DLGG STQ+T+ + +E E + +G R++THSFL G + RL
Sbjct 204 AAFDLGGGSTQLTYWPNNEAVFSEHVGYERDIDFFGHHIRLFTHSFL--GNGLIAARLNI 261
Query 60 RALQT----HGSH----PCWPRGY 75
L+T +H C P GY
Sbjct 262 LQLETDNEIESTHQLITSCMPEGY 285
> Hs4557423
Length=484
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 5 LDLGGASTQITF----ETTSPTEDPANEVQLRLYGQRYRVYTHSFLCYG 49
LDLGG STQI F E T P LR++ + Y++Y++S+L G
Sbjct 250 LDLGGGSTQIAFLPRVEGTLQASPPGYLTALRMFNRTYKLYSYSYLGLG 298
> At4g19180_2
Length=521
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/129 (24%), Positives = 51/129 (39%), Gaps = 21/129 (16%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLARALQT 64
LDLGG+S Q+TFE T + N + LR+ + + +S YG + R + L+
Sbjct 158 LDLGGSSLQVTFENEERTHNETN-LNLRIGSVNHHLSAYSLAGYGLNDAFDRSVVHLLKK 216
Query 65 --------------HGSHPCWPRGYSTHVMLWDVYESPC-TAAQRPQAFNRSTRVSLAGS 109
HPC GY+ + S C ++ Q + + L G+
Sbjct 217 LPNVNKSDLIEGKLEMKHPCLNSGYNGQYIC-----SQCASSVQGGKKGKSGVSIKLVGA 271
Query 110 SDPALCRSL 118
+ C +L
Sbjct 272 PNWGECSAL 280
> At3g04080
Length=471
Score = 35.8 bits (81), Expect = 0.027, Method: Composition-based stats.
Identities = 25/87 (28%), Positives = 41/87 (47%), Gaps = 23/87 (26%)
Query 5 LDLGGASTQITFETTSPTEDPANEV-----------QLRLYGQRYRVYTHSFLCYG---- 49
+DLGG S Q+ + P ED A ++ L G++Y +Y HS+L YG
Sbjct 220 VDLGGGSVQMAYAI--PEEDAATAPKPVEGEDSYVREMYLKGRKYFLYVHSYLHYGLLAA 277
Query 50 RDQVLRRLLARALQTHGSHPCWPRGYS 76
R ++L+ + ++PC GY+
Sbjct 278 RAEILK------VSEDSNNPCIATGYA 298
> At5g21040
Length=559
Score = 34.3 bits (77), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 50/113 (44%), Gaps = 13/113 (11%)
Query 4 PLDLGGASTQITFETTSPTEDPANEVQLRLYGQRYRVYTHSFLCYGRDQVLRRLLARALQ 63
P+ G AS+ ++ E + E + R + RY + T YG + +R + A
Sbjct 139 PVVFGAASSGLSDERSWKDLFVEREFRSRTFLGRYSIDT----LYGHTEAVRTVFLLA-- 192
Query 64 THGSHPCWPRGYSTHVMLWDVYESPCTAAQRP-----QAFNRSTRVSLAGSSD 111
+ + GY + V +WD+ E AA +P +A T++ +AG +D
Sbjct 193 --SAKLVFTSGYDSIVRMWDMEEGLSIAASKPLGCTIRALAADTKLLVAGGTD 243
> At5g18280
Length=472
Score = 33.5 bits (75), Expect = 0.12, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 19/84 (22%)
Query 5 LDLGGASTQITFETTS--------PTEDPANEV-QLRLYGQRYRVYTHSFLCYG----RD 51
+DLGG S Q+ + + P E + V ++ L G++Y +Y HS+L YG R
Sbjct 221 VDLGGGSVQMAYAISEEDAASAPKPLEGEDSYVREMYLKGRKYFLYVHSYLHYGLLAARA 280
Query 52 QVLRRLLARALQTHGSHPCWPRGY 75
++L+ + +PC GY
Sbjct 281 EILK------VSEDSENPCIVAGY 298
> At1g49700
Length=245
Score = 32.0 bits (71), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 4/69 (5%)
Query 58 LARALQTHGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCRS 117
L + L+T+G + T + L D+ ++P T Q+P+ ++S + GS+D A
Sbjct 88 LEQWLETNG----FANIEDTFLCLSDLVDAPVTEVQKPKDKDQSCDQWINGSNDLASIEE 143
Query 118 LVSQLFNSS 126
VS L N S
Sbjct 144 TVSSLINPS 152
> Hs10047120
Length=406
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 4/50 (8%)
Query 69 PCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCRSL 118
P P G + L D++ +P TAA Q N ST S DP+L RS+
Sbjct 266 PPAPSGRAYTSPLIDMFNNPATAAPNSQRVNNST----GTSEDPSLQRSV 311
> CE28748
Length=552
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 17/61 (27%), Positives = 31/61 (50%), Gaps = 11/61 (18%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQLRLYGQ---------RYRVYTHSFLCYGRDQVLR 55
+D+GGAS QI FE P D + + + +Y+++ +FL YG ++ +R
Sbjct 213 IDMGGASAQIAFEL--PDTDSFSSINVENINLGCREDDSLFKYKLFVTTFLGYGVNEGIR 270
Query 56 R 56
+
Sbjct 271 K 271
> At3g19210
Length=688
Score = 29.3 bits (64), Expect = 2.4, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 37/62 (59%), Gaps = 9/62 (14%)
Query 5 LDLGGASTQITFETTSPTEDPANEVQL--RLY--GQRYRVYTHSFLCYG--RDQVLRRLL 58
L+L GA+ + F+ P +PAN+ Q R++ GQ+ RVY + FL G ++V +R +
Sbjct 514 LNLIGANRLVLFD---PDWNPANDKQAAARVWRDGQKKRVYVYRFLSTGTIEEKVYQRQM 570
Query 59 AR 60
++
Sbjct 571 SK 572
> YGR067c
Length=794
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 34/74 (45%), Gaps = 11/74 (14%)
Query 38 YRVYTHSFLCYGRDQVLRRLLARALQTHGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQA 97
Y + H+ + RD +L L + + T +L D+ C A Q+P
Sbjct 635 YHILLHAIVSSLRDNILYPYLIYSPHLNDQ---------TKALLTDISNWSCFALQQP-- 683
Query 98 FNRSTRVSLAGSSD 111
F +++R SL+G+SD
Sbjct 684 FRKTSRGSLSGASD 697
> SPAC23E2.03c
Length=569
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 2/34 (5%)
Query 13 QITFETTSPTEDPANEVQLRLYGQRYRVYTHSFL 46
+I FET + T + ++QLR+ G Y VY H FL
Sbjct 21 KICFETNAATSPRSFQIQLRIKG--YAVYHHGFL 52
> Hs7019465
Length=376
Score = 28.1 bits (61), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 78 HVMLWDVYESPCTAAQRPQAFNRSTRVSLAGSSDPALCRSL 118
H M+ ++Y PC A + Q T G +DP + R L
Sbjct 296 HDMVTEMYSGPCVAMEIQQNNATKTFREFCGPADPEIARHL 336
> Hs22058533
Length=356
Score = 27.3 bits (59), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 28/67 (41%), Gaps = 0/67 (0%)
Query 44 SFLCYGRDQVLRRLLARALQTHGSHPCWPRGYSTHVMLWDVYESPCTAAQRPQAFNRSTR 103
SF C G++ + R+LQ P R V W+ YE T + +NR
Sbjct 206 SFSCLGKNLKQQHREFRSLQKQVHPPTVRRREQGPVWFWNRYEDGDTTQLMSRGWNRFGS 265
Query 104 VSLAGSS 110
+ L+G S
Sbjct 266 MPLSGHS 272
Lambda K H
0.322 0.133 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40