bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4425_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
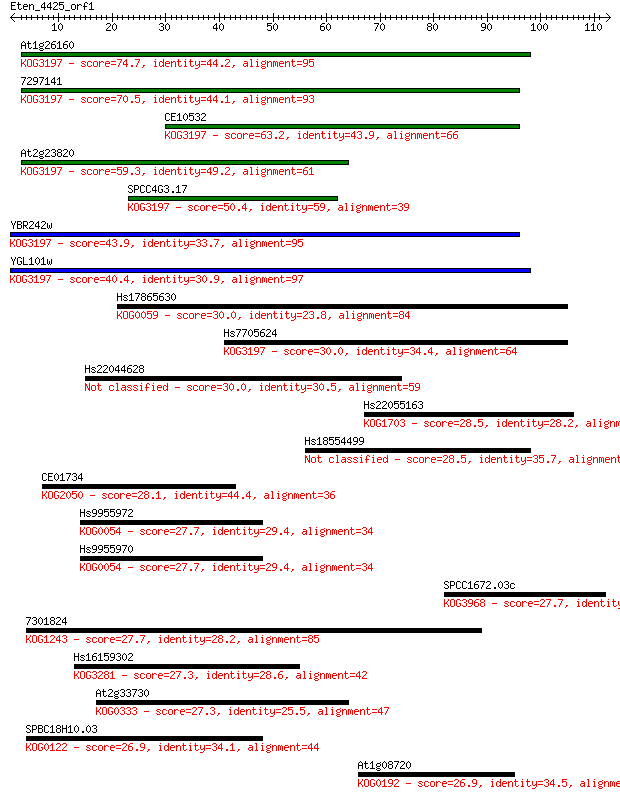
At1g26160 74.7 4e-14
7297141 70.5 7e-13
CE10532 63.2 1e-10
At2g23820 59.3 2e-09
SPCC4G3.17 50.4 7e-07
YBR242w 43.9 7e-05
YGL101w 40.4 8e-04
Hs17865630 30.0 0.97
Hs7705624 30.0 1.0
Hs22044628 30.0 1.1
Hs22055163 28.5 2.9
Hs18554499 28.5 3.5
CE01734 28.1 4.3
Hs9955972 27.7 4.8
Hs9955970 27.7 4.8
SPCC1672.03c 27.7 5.4
7301824 27.7 5.8
Hs16159302 27.3 7.6
At2g33730 27.3 7.9
SPBC18H10.03 26.9 8.2
At1g08720 26.9 8.6
> At1g26160
Length=248
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 56/96 (58%), Gaps = 1/96 (1%)
Query 3 EKREKEKNAFLFICKFLDDK-RAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYE 61
EK +E A +C+ L RA+E+ ELW EYE + EA +VKD DK +MI +A EYE
Sbjct 146 EKSRRETAALKEMCEVLGGGLRAEEITELWLEYENNASLEANIVKDFDKVEMILQALEYE 205
Query 62 NSHGVCLEEFFSSTENAFRTEVFKELNRSLRQQRSS 97
HG L+EFF ST F+TE+ K + +R S
Sbjct 206 AEHGKVLDEFFISTAGKFQTEIGKSWAAEINARRKS 241
> 7297141
Length=388
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 55/95 (57%), Gaps = 3/95 (3%)
Query 3 EKREKEKNAFLFICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYEN 62
+KR E A ICK ++ R K + EL+EEYE G+T E+ VKD D+ DM+ +AFEYE
Sbjct 261 DKRAMEFKAMEDICKLIE-PRGKRIMELFEEYEHGQTAESKFVKDLDRLDMVMQAFEYEK 319
Query 63 SHGVCL--EEFFSSTENAFRTEVFKELNRSLRQQR 95
L +EFF STE F K+L + +QR
Sbjct 320 RDNCLLKHQEFFDSTEGKFNHPFVKKLVNEIYEQR 354
> CE10532
Length=158
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 30 LWEEYERGETPEAMLVKDADKFDMITKAFEYENSHGVCLEEFFSSTENAFRTEVFKELNR 89
LW+EYE + A +VK DKFDMI +A +YE +H + L++FF+ST + E F +R
Sbjct 85 LWKEYEEASSLTARVVKHLDKFDMIVQADKYEKTHEIDLQQFFTSTVGVLKMEPFATWDR 144
Query 90 SLRQQR 95
LR+ R
Sbjct 145 ELRENR 150
> At2g23820
Length=243
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/62 (48%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 3 EKREKEKNAFLFICKFLDD-KRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYE 61
EK +E A +CK L +RAKE+ ELW EYE +PEA +VKD DK ++I +A EYE
Sbjct 160 EKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSSPEAKVVKDFDKVELILQALEYE 219
Query 62 NS 63
Sbjct 220 QG 221
> SPCC4G3.17
Length=198
Score = 50.4 bits (119), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 23/39 (58%), Positives = 30/39 (76%), Gaps = 0/39 (0%)
Query 23 RAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYE 61
+A+E++EL+ EYE TPEA VKD DKF+MI + FEYE
Sbjct 113 QAEEIKELFLEYESASTPEAKFVKDIDKFEMIAQMFEYE 151
> YBR242w
Length=238
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 51/99 (51%), Gaps = 5/99 (5%)
Query 1 EKEKREKEKNAFL--FICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAF 58
EK +RE E +L + K ++ AKE+ + W YE + EA VKD DK++M+ + F
Sbjct 128 EKHRREWETIKYLCNALIKPYNEIAAKEIMDDWLAYENVTSLEARYVKDIDKYEMLVQCF 187
Query 59 EYENSHGVC--LEEFFSSTENAFRTEVFKELNRSLRQQR 95
EYE + ++FF + + +T+ K L QR
Sbjct 188 EYEREYKGTKNFDDFFGAVA-SIKTDEVKGWTSDLVVQR 225
> YGL101w
Length=215
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 53/101 (52%), Gaps = 5/101 (4%)
Query 1 EKEKREKEKNAFL--FICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAF 58
EK +RE E +L I + + ++E+ + W YE+ E VKD DK++M+ + F
Sbjct 109 EKHRREFETVKYLCESIIRPCSESASREILDDWLAYEKQTCLEGRYVKDIDKYEMLVQCF 168
Query 59 EYENSHGV--CLEEFFSSTENAFRTEVFKELNRSLRQQRSS 97
EYE + L++F + N +T+ K+ +SL + R +
Sbjct 169 EYEQKYNGKKDLKQFLGAI-NDIKTDEVKKWTQSLLEDRQA 208
> Hs17865630
Length=1642
Score = 30.0 bits (66), Expect = 0.97, Method: Composition-based stats.
Identities = 20/88 (22%), Positives = 44/88 (50%), Gaps = 6/88 (6%)
Query 21 DKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEY----ENSHGVCLEEFFSSTE 76
D+ +E++ ++ R E+ ++L K D+ + + + E H +EE+ S
Sbjct 1544 DRLQREIQYIFPNASRQESFSSILAYKIPKEDVQSLSQSFFKLEEAKHAFAIEEY--SFS 1601
Query 77 NAFRTEVFKELNRSLRQQRSSCCSISSS 104
A +VF EL + ++ +SC +++S+
Sbjct 1602 QATLEQVFVELTKEQEEEDNSCGTLNST 1629
> Hs7705624
Length=170
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 41 EAMLVKDADKFDMITKAFEYEN-SHGVC-LEEFFSSTENAFRTEVFKELNRSLRQQRSSC 98
EA VK D+ +MI +A EYE+ H L++F+ ST F +L L +RS+
Sbjct 101 EAKFVKQLDQCEMILQASEYEDLEHKPGRLQDFYDSTAGKFNHPEIVQLVSELEAERSTN 160
Query 99 CSISSS 104
+ ++S
Sbjct 161 IAAAAS 166
> Hs22044628
Length=1395
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 15 ICKFLDDKRAKEVRELWEEYERGETPEAMLVKDA-DKFDMITKAFEYENSHGVCLEEFFS 73
+C L D ++K+ ++ ++ E +PEA L A D+ +M +AF + VCL+E +
Sbjct 134 LCSRLKDLQSKQEEKIHKKLEGSPSPEAELSPPAKDQVEMYYEAFPPLSEKPVCLQEIMT 193
> Hs22055163
Length=727
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 11/39 (28%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 67 CLEEFFSSTENAFRTEVFKELNRSLRQQRSSCCSISSSC 105
C E+FF+ T++ E+ +LRQ + C + ++C
Sbjct 601 CYEQFFAPLCAKCNTKIMGEVMHALRQTWHTTCFVCAAC 639
> Hs18554499
Length=1142
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 56 KAFEYENSHGVCLEEFFSSTENAFRTEVFKELNRSLRQQRSS 97
KAFE EN + ++FS+ +F T+ +E N R ++SS
Sbjct 440 KAFEEENQTIRRMAKYFSTPNKSFHTQYGEEENCHPRGEKSS 481
> CE01734
Length=766
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 3/39 (7%)
Query 7 KEKNAFLFICKF---LDDKRAKEVRELWEEYERGETPEA 42
KE+ AFL K + RA++ +ELWE+ G+TP+A
Sbjct 227 KERKAFLKELKLKRKPEGARAQKCKELWEKIRMGKTPKA 265
> Hs9955972
Length=1238
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 10/34 (29%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 14 FICKFLDDKRAKEVRELWEEYERGETPEAMLVKD 47
F+C + D+ + + W E E EA+L++D
Sbjct 848 FLCNYAPDEDQGHLEDSWTALEGAEDKEALLIED 881
> Hs9955970
Length=1527
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 10/34 (29%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 14 FICKFLDDKRAKEVRELWEEYERGETPEAMLVKD 47
F+C + D+ + + W E E EA+L++D
Sbjct 848 FLCNYAPDEDQGHLEDSWTALEGAEDKEALLIED 881
> SPCC1672.03c
Length=527
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 18/33 (54%), Gaps = 3/33 (9%)
Query 82 EVFKELNRSLRQQRSSCCSISSSCC---CCVQK 111
+VFKEL ++ R+ C SCC CC ++
Sbjct 469 QVFKELTQAHLLPRTQCVDTPPSCCGGHCCKEE 501
> 7301824
Length=873
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 42/91 (46%), Gaps = 7/91 (7%)
Query 4 KREKEKNAFL-----FICKFLDDK-RAKEVRELWEEYERGETPEAMLVKDADKFDMITKA 57
K + EKN F + F D+ R K + +L YE G+ A+L K +
Sbjct 285 KDKAEKNRFFSGLTTHLDNFPDNVCRHKILPQLITAYEYGDAGSAVLAP-MFKLGKLLDE 343
Query 58 FEYENSHGVCLEEFFSSTENAFRTEVFKELN 88
EY+ C+ + F+ST+ R+ + ++L+
Sbjct 344 VEYQKRIVPCVVKLFASTDRVTRSRLLQQLD 374
> Hs16159302
Length=328
Score = 27.3 bits (59), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 13 LFICKFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMI 54
+F + + +K A+E++++W++Y + V A+KFD+I
Sbjct 147 IFNIEMVKEKTAEEIKQIWQQYFAAKD-TVYAVIPAEKFDLI 187
> At2g33730
Length=733
Score = 27.3 bits (59), Expect = 7.9, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 30/47 (63%), Gaps = 3/47 (6%)
Query 17 KFLDDKRAKEVRELWEEYERGETPEAMLVKDADKFDMITKAFEYENS 63
K ++ ++ KE+ + E+Y G+ P+ +++ ++KF +F++EN+
Sbjct 150 KLVEREKEKELDAIKEQYLGGKKPKKRVIRPSEKFRF---SFDWENT 193
> SPBC18H10.03
Length=282
Score = 26.9 bits (58), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Query 4 KREKEKNAFLFICKFLDDKRAKEVRELWEEYERGETPEAMLVKD 47
KRE++ +A L + DD R +E+R+L+ + G L KD
Sbjct 195 KRERDDSATLRVTNLSDDTREEELRDLFRRF--GGIQRVYLAKD 236
> At1g08720
Length=1015
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 66 VCLEEFFSSTENAFRTEVFKELNRSLRQQ 94
V + F+STEN FR + +++NR+ +
Sbjct 646 VAVPSSFTSTENQFRPSIVEDMNRNTNNE 674
Lambda K H
0.319 0.131 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1188972946
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40