bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
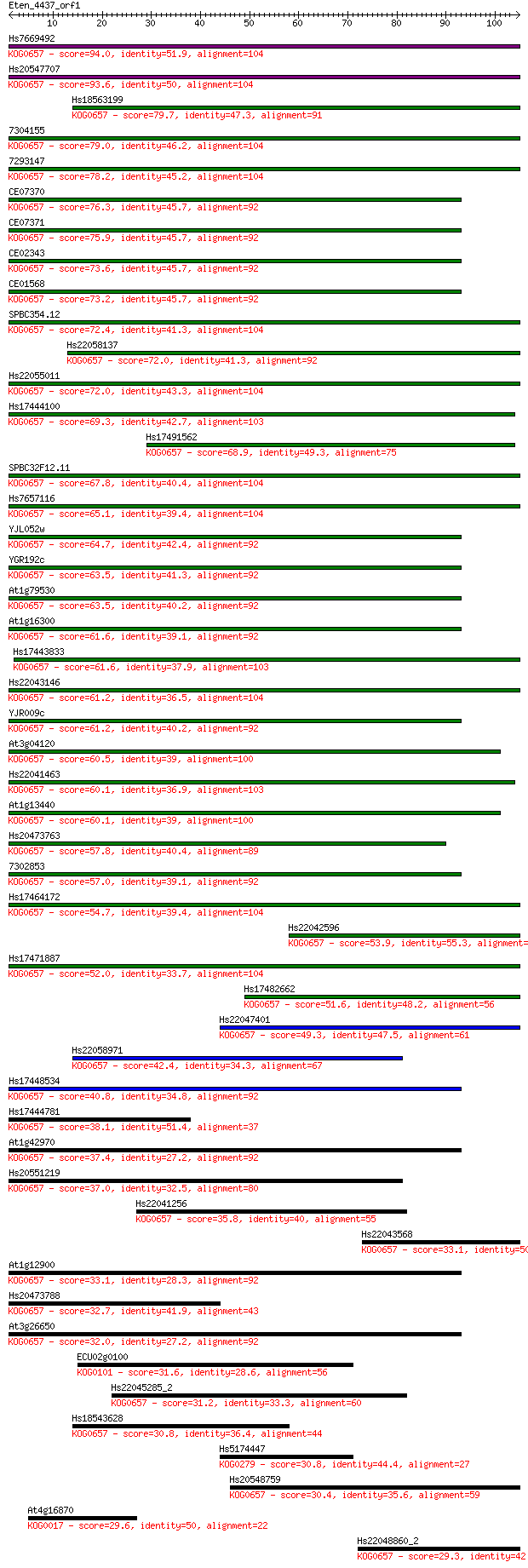
Query= Eten_4437_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
Hs7669492 94.0 5e-20
Hs20547707 93.6 8e-20
Hs18563199 79.7 1e-15
7304155 79.0 2e-15
7293147 78.2 4e-15
CE07370 76.3 1e-14
CE07371 75.9 2e-14
CE02343 73.6 8e-14
CE01568 73.2 1e-13
SPBC354.12 72.4 2e-13
Hs22058137 72.0 2e-13
Hs22055011 72.0 3e-13
Hs17444100 69.3 2e-12
Hs17491562 68.9 2e-12
SPBC32F12.11 67.8 4e-12
Hs7657116 65.1 3e-11
YJL052w 64.7 4e-11
YGR192c 63.5 8e-11
At1g79530 63.5 8e-11
At1g16300 61.6 3e-10
Hs17443833 61.6 4e-10
Hs22043146 61.2 4e-10
YJR009c 61.2 5e-10
At3g04120 60.5 8e-10
Hs22041463 60.1 1e-09
At1g13440 60.1 1e-09
Hs20473763 57.8 5e-09
7302853 57.0 7e-09
Hs17464172 54.7 4e-08
Hs22042596 53.9 6e-08
Hs17471887 52.0 2e-07
Hs17482662 51.6 3e-07
Hs22047401 49.3 2e-06
Hs22058971 42.4 2e-04
Hs17448534 40.8 6e-04
Hs17444781 38.1 0.004
At1g42970 37.4 0.008
Hs20551219 37.0 0.009
Hs22041256 35.8 0.019
Hs22043568 33.1 0.11
At1g12900 33.1 0.14
Hs20473788 32.7 0.19
At3g26650 32.0 0.26
ECU02g0100 31.6 0.38
Hs22045285_2 31.2 0.52
Hs18543628 30.8 0.62
Hs5174447 30.8 0.65
Hs20548759 30.4 0.91
At4g16870 29.6 1.5
Hs22048860_2 29.3 1.9
> Hs7669492
Length=335
Score = 94.0 bits (232), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/104 (51%), Positives = 60/104 (57%), Gaps = 0/104 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPT V V L E P + + KK+V QASEG KG LG +V S D NSDTH
Sbjct 232 AFRVPTANVSVVDLTCRLEKPAKYDDIKKVVKQASEGPLKGILGYTEHQVVSSDFNSDTH 291
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
T D GA I LN FVK S YDN FG +NR LM M SK+
Sbjct 292 SSTFDAGAGIALNDHFVKLISWYDNEFGYSNRVVDLMAHMASKE 335
> Hs20547707
Length=303
Score = 93.6 bits (231), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 52/104 (50%), Positives = 58/104 (55%), Gaps = 0/104 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
A RVPT V V L E P + + KK+V QASEG KG LG G +V S D NSDTH
Sbjct 200 ALRVPTANVSVVDLTYHLEKPAKYDDIKKVVKQASEGPLKGILGYTGHQVVSSDFNSDTH 259
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
T + GA LN FVK S YDN FG NR LM M SK+
Sbjct 260 SSTFNAGAGTALNDHFVKLISWYDNEFGYNNRVVDLMAHMASKE 303
> Hs18563199
Length=243
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 43/91 (47%), Positives = 52/91 (57%), Gaps = 2/91 (2%)
Query 14 LPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTHFFTLDGGAAIPLN 73
LPS + P + + KK++ QASE KG LG ++ S D NSDT T D GA I LN
Sbjct 155 LPSGK--PAKYYDIKKMMKQASEDPIKGILGYTEHQIVSSDFNSDTQSSTFDAGAGITLN 212
Query 74 APFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
FVK S YDN FG +NR LM M SK+
Sbjct 213 DHFVKLISWYDNEFGYSNRVVDLMAHMASKE 243
> 7304155
Length=331
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/104 (46%), Positives = 53/104 (50%), Gaps = 0/104 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L + E K V +AS+G KG LG V S D SDTH
Sbjct 228 AFRVPTPNVSVVDLTVRLGKGATYDEIKAKVEEASKGPLKGILGYTDEEVVSTDFFSDTH 287
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
D A I LN FVK S YDN FG +NR L+ M SKD
Sbjct 288 SSVFDAKAGISLNDKFVKLISWYDNEFGYSNRVIDLIKYMQSKD 331
> 7293147
Length=332
Score = 78.2 bits (191), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 47/104 (45%), Positives = 53/104 (50%), Gaps = 0/104 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L ++ E K V +A+ G KG LG V S D SDTH
Sbjct 229 AFRVPTPNVSVVDLTVRLGKGASYDEIKAKVQEAANGPLKGILGYTDEEVVSTDFLSDTH 288
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
D A I LN FVK S YDN FG +NR L+ M SKD
Sbjct 289 SSVFDAKAGISLNDKFVKLISWYDNEFGYSNRVIDLIKYMQSKD 332
> CE07370
Length=341
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 42/92 (45%), Positives = 53/92 (57%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L + E P + + KK++ A++G KG L +V S D SDT+
Sbjct 238 AFRVPTPDVSVVDLTARLEKPASLDDIKKVIKAAADGPMKGILAYTEDQVVSTDFVSDTN 297
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D GA+I LN FVK S YDN FG +NR
Sbjct 298 SSIFDAGASISLNPHFVKLVSWYDNEFGYSNR 329
> CE07371
Length=341
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 42/92 (45%), Positives = 53/92 (57%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L + E P + + KK++ A++G KG L +V S D SDT+
Sbjct 238 AFRVPTPDVSVVDLTARLEKPASLDDIKKVIKAAADGPMKGILAYTEDQVVSTDFVSDTN 297
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D GA+I LN FVK S YDN FG +NR
Sbjct 298 SSIFDAGASISLNPHFVKLVSWYDNEFGYSNR 329
> CE02343
Length=341
Score = 73.6 bits (179), Expect = 8e-14, Method: Composition-based stats.
Identities = 42/92 (45%), Positives = 50/92 (54%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L E P + + KK+V A++G KG L +V S D SD H
Sbjct 238 AFRVPTPDVSVVDLTVRLEKPASMDDIKKVVKAAADGPMKGILAYTEDQVVSTDFVSDPH 297
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D GA I LN FVK S YDN +G +NR
Sbjct 298 SSIFDAGACISLNPNFVKLVSWYDNEYGYSNR 329
> CE01568
Length=341
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 42/92 (45%), Positives = 50/92 (54%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L E P + + KK+V A++G KG L +V S D SD H
Sbjct 238 AFRVPTPDVSVVDLTVRLEKPASMDDIKKVVKAAADGPMKGILAYTEDQVVSTDFVSDPH 297
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D GA I LN FVK S YDN +G +NR
Sbjct 298 SSIFDTGACISLNPNFVKLVSWYDNEYGYSNR 329
> SPBC354.12
Length=335
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 43/104 (41%), Positives = 51/104 (49%), Gaps = 0/104 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L P N+ + K + ASEG KG LG V S D D H
Sbjct 232 AFRVPTPDVSVVDLTVKLAKPTNYEDIKAAIKAASEGPMKGVLGYTEDSVVSTDFCGDNH 291
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
D A I L+ FVK S YDN +G ++R L+ SKD
Sbjct 292 SSIFDASAGIQLSPQFVKLVSWYDNEWGYSHRVVDLVAYTASKD 335
> Hs22058137
Length=307
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 50/92 (54%), Gaps = 5/92 (5%)
Query 13 VLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTHFFTLDGGAAIPL 72
V+ E P + + KK+ EG KG LG +V S+D NS+T + D G + L
Sbjct 221 VMDINHEKPGKYDDIKKV-----EGPLKGILGYNEHQVVSFDFNSNTQYSIFDAGVGVTL 275
Query 73 NAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
N FVK SCYDN FG +NR LMV + SK+
Sbjct 276 NDHFVKLISCYDNEFGCSNRVVDLMVHIASKE 307
> Hs22055011
Length=288
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 58/106 (54%), Gaps = 2/106 (1%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AF V T K+ V L + +K+V QASEG KG LG +V S + NS+TH
Sbjct 183 AFCVSTAKLSVVDLTCCLGKTAKYDGIEKVVGQASEGPLKGILGYTEHQVVSSEFNSNTH 242
Query 61 --FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
F ++ G I LN FVK S YDN FG++NR LMV + SK+
Sbjct 243 SSAFNVNEGNDIALNNHFVKLISWYDNEFGNSNRVIDLMVHLTSKE 288
> Hs17444100
Length=324
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 44/103 (42%), Positives = 54/103 (52%), Gaps = 0/103 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AF V T KV V L E P + + KK+V Q SEG KG LG + S + NS+TH
Sbjct 210 AFCVTTAKVSVINLTCHLEKPAKYDDIKKVVKQTSEGPLKGILGYTEHQAVSSNFNSNTH 269
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSK 103
T + A I LN FVK S Y + FG +N+ LM M SK
Sbjct 270 SSTFNVRAGIALNNHFVKIISWYSDEFGYSNKVVDLMAHMASK 312
> Hs17491562
Length=242
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/75 (49%), Positives = 43/75 (57%), Gaps = 0/75 (0%)
Query 29 KLVNQASEGLFKGFLGSPGARVFSWDLNSDTHFFTLDGGAAIPLNAPFVKFFSCYDNNFG 88
K+V QASEG +G LG +V + D NS TH T G I LN FVK S Y+N FG
Sbjct 165 KVVKQASEGTLEGILGYTEHQVVASDFNSITHSSTFKAGVGIALNNHFVKLISWYENEFG 224
Query 89 STNRGGALMVPMPSK 103
+NR LMV M SK
Sbjct 225 YSNRVVDLMVHMASK 239
> SPBC32F12.11
Length=336
Score = 67.8 bits (164), Expect = 4e-12, Method: Composition-based stats.
Identities = 42/104 (40%), Positives = 50/104 (48%), Gaps = 0/104 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L P N+ + K + ASEG KG LG V S D D H
Sbjct 232 AFRVPTPDVSVVDLTVKLAKPTNYEDIKAAIKAASEGPMKGVLGYTEDAVVSTDFCGDNH 291
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
D A I L+ FVK S YDN +G + R L+ +KD
Sbjct 292 SSIFDASAGIQLSPQFVKLVSWYDNEWGYSRRVVDLVAYTAAKD 335
> Hs7657116
Length=408
Score = 65.1 bits (157), Expect = 3e-11, Method: Composition-based stats.
Identities = 41/104 (39%), Positives = 50/104 (48%), Gaps = 0/104 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L P + K+ V A++G G L V S D DTH
Sbjct 304 AFRVPTPDVSVVDLTCRLAQPAPYSAIKEAVKAAAKGPMAGILAYTEDEVVSTDFLGDTH 363
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
D A I LN FVK S YDN +G ++R L+ M S+D
Sbjct 364 SSIFDAKAGIALNDNFVKLISWYDNEYGYSHRVVDLLRYMFSRD 407
> YJL052w
Length=332
Score = 64.7 bits (156), Expect = 4e-11, Method: Composition-based stats.
Identities = 39/92 (42%), Positives = 45/92 (48%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPT V V L E + + KK V A+EG KG LG V S D DTH
Sbjct 230 AFRVPTVDVSVVDLTVKLEKEATYDQIKKAVKAAAEGPMKGVLGYTEDAVVSSDFLGDTH 289
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D A I L+ FVK S YDN +G + R
Sbjct 290 ASIFDASAGIQLSPKFVKLISWYDNEYGYSAR 321
> YGR192c
Length=332
Score = 63.5 bits (153), Expect = 8e-11, Method: Composition-based stats.
Identities = 38/92 (41%), Positives = 45/92 (48%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPT V V L + E KK+V A+EG KG LG V S D D+H
Sbjct 230 AFRVPTVDVSVVDLTVKLNKETTYDEIKKVVKAAAEGKLKGVLGYTEDAVVSSDFLGDSH 289
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D A I L+ FVK S YDN +G + R
Sbjct 290 SSIFDASAGIQLSPKFVKLVSWYDNEYGYSTR 321
> At1g79530
Length=422
Score = 63.5 bits (153), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 45/92 (48%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPT V V L E ++ + K + ASEG KG LG V S D D+
Sbjct 316 AFRVPTSNVSVVDLTCRLEKGASYEDVKAAIKHASEGPLKGILGYTDEDVVSNDFVGDSR 375
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D A I L+ FVK S YDN +G +NR
Sbjct 376 SSIFDANAGIGLSKSFVKLVSWYDNEWGYSNR 407
> At1g16300
Length=420
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 46/92 (50%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPTP V V L E ++ + K + ASEG +G LG V S D D+
Sbjct 314 AFRVPTPNVSVVDLTCRLEKDASYEDVKAAIKFASEGPLRGILGYTEEDVVSNDFLGDSR 373
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D A I L+ F+K S YDN +G +NR
Sbjct 374 SSIFDANAGIGLSKSFMKLVSWYDNEWGYSNR 405
> Hs17443833
Length=268
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 45/103 (43%), Gaps = 11/103 (10%)
Query 2 FRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTHF 61
F V V V L E P + + KK G G +V S D N DTH
Sbjct 177 FCVHITNVSVMDLTCRLEKPAKYDDIKK-----------GIWGDTEHQVVSSDFNRDTHS 225
Query 62 FTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
T D GA I L FVK S YDN FG +NR LM + SK+
Sbjct 226 STFDAGAGIALKDHFVKLISWYDNEFGYSNRVVDLMAHIASKE 268
> Hs22043146
Length=175
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/104 (36%), Positives = 47/104 (45%), Gaps = 11/104 (10%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPT V V L ++ P + + KK G LG +V S + S H
Sbjct 83 AFRVPTTNVLVMNLTCHQKIPAKYDDIKK-----------GILGYNEHQVVSSNFISIMH 131
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
F T D G I LN F+K S YDN F NR L+ M K+
Sbjct 132 FSTFDAGTGIALNDHFIKLISWYDNEFDYNNRVVNLIAHMAFKE 175
> YJR009c
Length=332
Score = 61.2 bits (147), Expect = 5e-10, Method: Composition-based stats.
Identities = 37/92 (40%), Positives = 45/92 (48%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVPT V V L + E KK+V A+EG KG LG V S D D++
Sbjct 230 AFRVPTVDVSVVDLTVKLNKETTYDEIKKVVKAAAEGKLKGVLGYTEDAVVSSDFLGDSN 289
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D A I L+ FVK S YDN +G + R
Sbjct 290 SSIFDAAAGIQLSPKFVKLVSWYDNEYGYSTR 321
> At3g04120
Length=338
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 39/100 (39%), Positives = 48/100 (48%), Gaps = 0/100 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
+FRVPT V V L E + E KK + + SEG KG LG V S D D
Sbjct 236 SFRVPTVDVSVVDLTVRLEKAATYDEIKKAIKEESEGKLKGILGYTEDDVVSTDFVGDNR 295
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPM 100
D A I L+ FVK S YDN +G ++R L+V M
Sbjct 296 SSIFDAKAGIALSDKFVKLVSWYDNEWGYSSRVVDLIVHM 335
> Hs22041463
Length=277
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 49/103 (47%), Gaps = 15/103 (14%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AF VPT V V L E P + + K++V + G GA SDTH
Sbjct 189 AFHVPTANVSVMDLTCSLEKPAKYDDIKEVVKRHRRG-------PSGA--------SDTH 233
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSK 103
+ T + GA I LN FV+ S YDN FG +N+ M+ M SK
Sbjct 234 YSTFNTGAGIALNGLFVRLVSGYDNEFGYSNKVITFMIHMASK 276
> At1g13440
Length=338
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 39/100 (39%), Positives = 48/100 (48%), Gaps = 0/100 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
+FRVPT V V L E + E KK + + SEG KG LG V S D D
Sbjct 236 SFRVPTVDVSVVDLTVRLEKAATYDEIKKAIKEESEGKMKGILGYTEDDVVSTDFVGDNR 295
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPM 100
D A I L+ FVK S YDN +G ++R L+V M
Sbjct 296 SSIFDAKAGIALSDKFVKLVSWYDNEWGYSSRVVDLIVHM 335
> Hs20473763
Length=219
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/89 (40%), Positives = 43/89 (48%), Gaps = 1/89 (1%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AF VPT + V L E P + KK+V QASEG KG L +V S L +DTH
Sbjct 82 AFHVPTTIMLVMDLTYHLEKPAKYDNIKKVVKQASEGPHKGILCYTENQVVS-SLTTDTH 140
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGS 89
TLD G I LN + FS N +
Sbjct 141 SSTLDPGTGIALNDHLLSSFSDMKMNLAT 169
> 7302853
Length=343
Score = 57.0 bits (136), Expect = 7e-09, Method: Composition-based stats.
Identities = 36/92 (39%), Positives = 40/92 (43%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AFRVP P V V L P + KK + AS+ KG LG V S D N
Sbjct 230 AFRVPVPNVSVVDLTCRLSKPAKMDDIKKCIKAASKCEMKGILGYVEEEVVSTDFNGSRF 289
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
D A I LN FVK S YDN G + R
Sbjct 290 ASVFDAKACIALNDNFVKLISWYDNETGYSCR 321
> Hs17464172
Length=322
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 41/105 (39%), Positives = 50/105 (47%), Gaps = 12/105 (11%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLG-SPGARVFSWDLNSDT 59
AF VPT V V L E P + G+ KG LG + +V S+D NSD
Sbjct 229 AFCVPTANVSVVDLIYHLEKPTKY-----------HGIKKGILGYNTEHQVVSFDFNSDI 277
Query 60 HFFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
H T D GA I LN VK S Y+N FG +NR +M SK+
Sbjct 278 HSSTFDVGAGIALNNHCVKLISWYENEFGYSNRVVDVMAHKSSKE 322
> Hs22042596
Length=140
Score = 53.9 bits (128), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 26/47 (55%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 58 DTHFFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
DTH T D GA I LN FVK SCYDN FG +NR M M SK+
Sbjct 94 DTHSSTFDTGAGIALNDHFVKLISCYDNEFGYSNRVVDFMAHMASKE 140
> Hs17471887
Length=198
Score = 52.0 bits (123), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 43/104 (41%), Gaps = 19/104 (18%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AF V T K+ V L E + + KK+V S D NS+ H
Sbjct 114 AFHVSTAKLLVMDLTCHLEKAAKYDDIKKVV-------------------VSSDFNSNFH 154
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
+ GA I LN FV S YDN FG +NR LM SK+
Sbjct 155 SSAFNAGAGIALNNHFVNLISWYDNEFGYSNRVVDLMAHKASKE 198
> Hs17482662
Length=325
Score = 51.6 bits (122), Expect = 3e-07, Method: Composition-based stats.
Identities = 27/56 (48%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 49 RVFSWDLNSDTHFFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
+V S D +SDTH T + GA I LN FVK S YD+ FG +N LM M SK+
Sbjct 270 QVVSSDFDSDTHSSTFNAGAGIALNNHFVKLISWYDDEFGYSNSMVDLMAHMASKE 325
> Hs22047401
Length=244
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Query 44 GSPG---ARVFSWDLNSDTHFF--TLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMV 98
G PG +V S D NSDTH + T + GA L+ FVK S YDN FG +NR L
Sbjct 179 GHPGLHEHQVVSSDFNSDTHTYSSTFNAGAGTALDGHFVKLVSWYDNEFGYSNRVVNLRA 238
Query 99 PMPSKD 104
M +K+
Sbjct 239 HMAAKE 244
> Hs22058971
Length=173
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 32/67 (47%), Gaps = 0/67 (0%)
Query 14 LPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTHFFTLDGGAAIPLN 73
L E + + K+++QA EG + L +V S D NSDTH D A+I L+
Sbjct 105 LTCILEKVYKYSDINKVLSQALEGPLEDTLAYAKDQVISCDFNSDTHSSIFDAWASIDLH 164
Query 74 APFVKFF 80
VKF
Sbjct 165 DHIVKFI 171
> Hs17448534
Length=400
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 32/92 (34%), Positives = 41/92 (44%), Gaps = 18/92 (19%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AF VP+ + V L E P + + K+V QASEGL K LG ++ H
Sbjct 141 AFHVPSANMLVRDLTWHLEKPAKYDDTNKVVKQASEGLLKDILGY-----------TEHH 189
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
AAI LN FVK S Y + +NR
Sbjct 190 -------AAIALNDYFVKLMSWYYSAVSYSNR 214
> Hs17444781
Length=218
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEG 37
AFRVPT V V L ENP + + KK+V QASE
Sbjct 174 AFRVPTTNVSVMHLTCHLENPAKYDDIKKVVKQASEA 210
> At1g42970
Length=447
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 40/93 (43%), Gaps = 1/93 (1%)
Query 1 AFRVPTPKV-FVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDT 59
A RVPTP V V ++ + + + + +A+ G KG L A + S D
Sbjct 314 ALRVPTPNVSVVDLVINVEKKGLTAEDVNEAFRKAANGPMKGILDVCDAPLVSVDFRCSD 373
Query 60 HFFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
T+D + + VK + YDN +G + R
Sbjct 374 VSTTIDSSLTMVMGDDMVKVVAWYDNEWGYSQR 406
> Hs20551219
Length=198
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 37/80 (46%), Gaps = 0/80 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
AF VPT + V L + P + KK++ +A +G + G +V S + NSD
Sbjct 118 AFCVPTANMLVVDLTCHIKKPAKYGNTKKVMKEARKGPSRTSWGYTEHQVVSSNFNSDID 177
Query 61 FFTLDGGAAIPLNAPFVKFF 80
T D GA I L+ VK
Sbjct 178 PSTFDVGAGIVLSDHSVKLI 197
> Hs22041256
Length=201
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 25/55 (45%), Gaps = 0/55 (0%)
Query 27 FKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTHFFTLDGGAAIPLNAPFVKFFS 81
KKLV QA + L +VF D NSDTH D A I L FVK +
Sbjct 134 IKKLVKQALGHFLRCILAYTEHQVFPSDFNSDTHPSGFDVRAGIALKDHFVKLIT 188
> Hs22043568
Length=247
Score = 33.1 bits (74), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 73 NAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
N FVK S +DN FG +NR MV M SK+
Sbjct 216 NDHFVKLISQHDNEFGYSNRVVGFMVHMASKE 247
> At1g12900
Length=399
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 26/92 (28%), Positives = 34/92 (36%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
A RVPTP V V L E A+E KG L + S D
Sbjct 295 ALRVPTPNVSVVDLVVQVSKKTFAEEVNAAFRDAAEKELKGILDVCDEPLVSVDFRCSDV 354
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
T+D + + VK + YDN +G + R
Sbjct 355 SSTIDSSLTMVMGDDMVKVIAWYDNEWGYSQR 386
> Hs20473788
Length=101
Score = 32.7 bits (73), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFL 43
AF VPT + V L E P + KK+V Q SE KG L
Sbjct 55 AFHVPTTNMLVMDLTCHLEKPAKY-NIKKVVKQTSEDPLKGIL 96
> At3g26650
Length=396
Score = 32.0 bits (71), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 34/92 (36%), Gaps = 0/92 (0%)
Query 1 AFRVPTPKVFVGVLPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTH 60
A RVPTP V V L E ++E KG L + S D
Sbjct 292 ALRVPTPNVSVVDLVVQVSKKTFAEEVNAAFRDSAEKELKGILDVCDEPLVSVDFRCSDF 351
Query 61 FFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNR 92
T+D + + VK + YDN +G + R
Sbjct 352 STTIDSSLTMVMGDDMVKVIAWYDNEWGYSQR 383
> ECU02g0100
Length=687
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query 15 PSPRENPPNFVEFKKLVN-QASEGLFKGFLGSPGARVFSWDLNSDTHFFTLDGGAAI 70
P E P+ V FKKL + ++ G PG ++++ + N++T+F+ G I
Sbjct 60 PGGEETYPSVVYFKKLPEGDGKKSKYEAVAGWPGLQMYNAEGNANTYFYAFKRGMGI 116
> Hs22045285_2
Length=246
Score = 31.2 bits (69), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 26/60 (43%), Gaps = 8/60 (13%)
Query 22 PNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNSDTHFFTLDGGAAIPLNAPFVKFFS 81
P + + KK+V ASE KG L S+ + F T + G I L FVK S
Sbjct 195 PKYDDIKKVVKLASEATLKGLL--------SYTEHQTPTFSTFNAGNGIALKDHFVKLIS 246
> Hs18543628
Length=251
Score = 30.8 bits (68), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 14 LPSPRENPPNFVEFKKLVNQASEGLFKGFLGSPGARVFSWDLNS 57
L ++ P + KK V QA + FKG LG +V S D N+
Sbjct 167 LTWHQDKPAKYDSIKKTVKQALDKTFKGILGFTEEQVVSSDFNA 210
> Hs5174447
Length=317
Score = 30.8 bits (68), Expect = 0.65, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 14/27 (51%), Gaps = 0/27 (0%)
Query 44 GSPGARVFSWDLNSDTHFFTLDGGAAI 70
G + WDLN H +TLDGG I
Sbjct 210 GGKDGQAMLWDLNEGKHLYTLDGGDII 236
> Hs20548759
Length=806
Score = 30.4 bits (67), Expect = 0.91, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 28/59 (47%), Gaps = 6/59 (10%)
Query 46 PGARVFSWDLNSDTHFFTLDGGAAIPLNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
P R+ W L+S T + I N FV+ S + FGS+NR LMV SK+
Sbjct 754 PDKRLTEWHLHSST----FNATTGITFNGHFVRLLSW--DEFGSSNRVVDLMVHTASKE 806
> At4g16870
Length=1474
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 5 PTPKVFVGVLPSPRENPPNFVE 26
PT + F+G LP+P NP N +E
Sbjct 875 PTHEAFIGPLPNPNRNPTNEIE 896
> Hs22048860_2
Length=238
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 72 LNAPFVKFFSCYDNNFGSTNRGGALMVPMPSKD 104
+ FV S YDN FG NR L+V M K+
Sbjct 206 ITTYFVNLISWYDNEFGYINRVLDLIVHMAFKE 238
Lambda K H
0.322 0.141 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40