bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4458_orf1
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
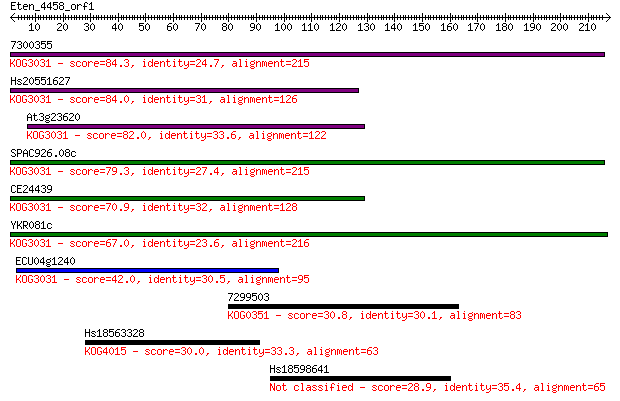
7300355 84.3 2e-16
Hs20551627 84.0 2e-16
At3g23620 82.0 7e-16
SPAC926.08c 79.3 5e-15
CE24439 70.9 2e-12
YKR081c 67.0 3e-11
ECU04g1240 42.0 8e-04
7299503 30.8 2.2
Hs18563328 30.0 3.4
Hs18598641 28.9 8.1
> 7300355
Length=289
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 53/215 (24%), Positives = 90/215 (41%), Gaps = 62/215 (28%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
F++ +E+L KN LF + S+SKKRP ++LGR+++N++LDM E + Y + F
Sbjct 40 FDDPSSLEFLTMKNDAALFTFGSTSKKRPDNIILGRIFENEVLDMFELGIKRYQAISEFK 99
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLI 120
+ A P L+ G W ++E+R RN+F D F+ +V + L G++ ++
Sbjct 100 N-EKIGACVKPCLVFNGPKWAQTEELRRLRNLFIDTFQREKVDS------IRLQGIEHVL 152
Query 121 AVSAVQATPGSAATAATTAAAAKAVIGGSEDCETEAPASGATTDGTDAPAADSGYSGDVL 180
+ + + D+
Sbjct 153 SFTV---------------------------------------------------TDDMN 161
Query 181 ICVRHYRIALVKTADAGGVGGPAVKLVEVGPQIDL 215
I +R YRI L K+ G P ++L E+GP D
Sbjct 162 ILMRSYRILLKKS----GQRTPRIELEEIGPSADF 192
> Hs20551627
Length=306
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 39/127 (30%), Positives = 71/127 (55%), Gaps = 9/127 (7%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+ +E+ ++K+ C LF + S +KKRP LV+GR+YD +LDM E + ++
Sbjct 72 FEDQTSLEFFSKKSDCSLFMFGSHNKKRPNNLVIGRMYDYHVLDMIELGIENFVSLKDIK 131
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPT-QKLYLGGVDRL 119
+ P G+ P+L+ G +DV+++ R +++ D FR GPT + L G++ +
Sbjct 132 NSKCP-EGTKPMLIFAGDDFDVTEDYRRLKSLLIDFFR-------GPTVSNIRLAGLEYV 183
Query 120 IAVSAVQ 126
+ +A+
Sbjct 184 LHFTALN 190
> At3g23620
Length=269
Score = 82.0 bits (201), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 67/124 (54%), Gaps = 8/124 (6%)
Query 7 IEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAAVQ--A 64
+E+ ++K C +F Y S +KKRP LVLGR+YD+Q+ D+ E + ++ AF+ + A
Sbjct 33 LEFFSQKTDCSIFVYGSHTKKRPDNLVLGRMYDHQVYDLIEVGIENFKSLRAFSYDKKFA 92
Query 65 PAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLIAVSA 124
P G+ P + G ++ E+++ + + DLFR V L L G+DR SA
Sbjct 93 PHEGTKPFICFIGEGFENVSELKHLKEVLTDLFRGEVV------DNLNLTGLDRAYVCSA 146
Query 125 VQAT 128
+ T
Sbjct 147 ISPT 150
> SPAC926.08c
Length=317
Score = 79.3 bits (194), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 59/215 (27%), Positives = 90/215 (41%), Gaps = 54/215 (25%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+A +E+ + KN L A+ +KKRP L R ++ ++LDM E + +Y +F+
Sbjct 69 FEDASSLEFFSEKNDAALAVMATHNKKRPHNLTWVRFFNYRVLDMIELGIVNYKSIQSFS 128
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLI 120
A G+ P++L QG ++D R+ +++F D FR P QKL G+ +I
Sbjct 129 ATPI-VPGTKPMILFQGPVFDAHPTYRHIKSLFLDFFRGE------PIQKLDSAGLSYVI 181
Query 121 AVSAVQATPGSAATAATTAAAAKAVIGGSEDCETEAPASGATTDGTDAPAADSGYSGDVL 180
VSA +A ED P L
Sbjct 182 VVSAAEA---------------------QEDETKPLP----------------------L 198
Query 181 ICVRHYRIALVKTADAGGVGGPAVKLVEVGPQIDL 215
+ R Y L+KT P V+L E+GP+ID
Sbjct 199 VHFRVYGTKLLKTK----TNLPRVELEEMGPRIDF 229
> CE24439
Length=297
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 64/128 (50%), Gaps = 7/128 (5%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFA 60
FE+ PI K LF S+SKK+P L GR YD Q+LDM E + Y ++ F
Sbjct 69 FEDETPIVRAGSKFDTSLFVLGSNSKKKPNCLTFGRTYDGQLLDMAELRITSYKSSSNFE 128
Query 61 AVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRLI 120
A + GS P ++L+G ++ +M+ N+ D FR +V + L G++ +I
Sbjct 129 AAKM-TLGSKPCVILEGAAFESDGDMKRIGNLMVDWFRGPKVDT------VRLEGLETVI 181
Query 121 AVSAVQAT 128
+A+ T
Sbjct 182 VFTALDET 189
> YKR081c
Length=344
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 51/217 (23%), Positives = 82/217 (37%), Gaps = 53/217 (24%)
Query 1 FENAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVA-HYTPAAAF 59
FE+ P+E+ + KN C L +SSKKR + R + +I DM E VA ++ + F
Sbjct 69 FEDMSPLEFFSEKNDCSLMVLMTSSKKRKNNMTFIRTFGYKIYDMIELMVADNFKLLSDF 128
Query 60 AAVQAPAAGSAPLLLLQGGLWDVSDEMRNARNIFGDLFRAAEVPPDGPTQKLYLGGVDRL 119
+ G P+ QG +D + +++F D FR
Sbjct 129 KKLTF-TVGLKPMFTFQGAAFDTHPVYKQIKSLFLDFFRG-------------------- 167
Query 120 IAVSAVQATPGSAATAATTAAAAKAVIGGSEDCETEAPASGATTDGTDAPAADSGYSGDV 179
+T A + VI + G DG P
Sbjct 168 ------------ESTDLQDVAGLQHVISMT--------IQGDFQDGEPLPN--------- 198
Query 180 LICVRHYRIALVKTADAGGVGGPAVKLVEVGPQIDLR 216
+ R Y++ K +D GG P ++LVE+GP++D +
Sbjct 199 -VLFRVYKLKSYK-SDQGGKRLPRIELVEIGPRLDFK 233
> ECU04g1240
Length=289
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 47/97 (48%), Gaps = 15/97 (15%)
Query 3 NAEPIEYLARKNGCGLFAYASSSKKRPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAAV 62
+ E IE L +K GLF S +K+R LV+GR ++++++DM EF + Y + F
Sbjct 95 SVEKIEKLMKKKRSGLFL--SVTKER--HLVIGRAFNDELIDMVEFKINRYLSVSDFECA 150
Query 63 QAPAAGSAPLLLLQGGLWDVSDEMRNAR--NIFGDLF 97
P ++LQ + N+R N+ D F
Sbjct 151 -GPELHMKYFVVLQN--------INNSRLENLVVDFF 178
> 7299503
Length=1487
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 25/86 (29%), Positives = 36/86 (41%), Gaps = 8/86 (9%)
Query 80 WDVSDEMRNARNIFGDLFRAAEV--PPDGPTQKLYLGG-VDRLIAVSAVQATPGSAATAA 136
WD +D R R + D F ++ D P LYLG + +L+ + TP
Sbjct 1200 WDKNDVHRLLRKMVIDGFLREDLIFTNDFPQAYLYLGNNISKLM-----EGTPNFEFAVT 1254
Query 137 TTAAAAKAVIGGSEDCETEAPASGAT 162
A AKA +G D T + A G +
Sbjct 1255 KNAKEAKAAVGSVSDGATSSTADGQS 1280
> Hs18563328
Length=279
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 29/63 (46%), Gaps = 5/63 (7%)
Query 28 RPARLVLGRVYDNQILDMQEFAVAHYTPAAAFAAVQAPAAGSAPLLLLQGGLWDVSDEMR 87
R RL+ R + ++ ++ A+AH PA+ AP GS PLL L D S E
Sbjct 205 RKVRLLAERTLNTEVPFLESLALAHSGPAST-----APGPGSLPLLPEFTHLVDRSSEHG 259
Query 88 NAR 90
R
Sbjct 260 TDR 262
> Hs18598641
Length=98
Score = 28.9 bits (63), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 28/70 (40%), Gaps = 5/70 (7%)
Query 95 DLFRAAEVPPDGPTQKLYLGGVDRLIAVSAVQATPGSAATAATTAAAAKAVIGGSEDC-- 152
+ R + PP G T +LG RL A A P S A ++ A GS C
Sbjct 13 PVLRCSWPPPTGRTGSHFLGLPRRLRCSPAPSAEPPSGPAAPSSRARGCRPCSGSRRCRA 72
Query 153 ---ETEAPAS 159
ET AP S
Sbjct 73 CAAETPAPLS 82
Lambda K H
0.316 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4040519078
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40