bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4473_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
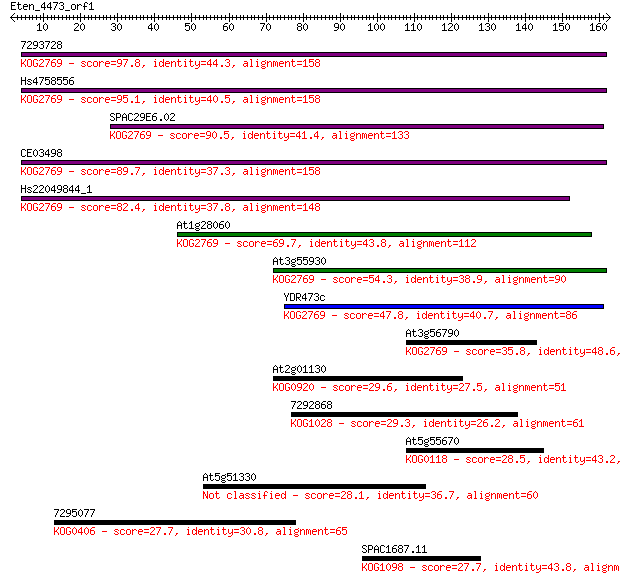
7293728 97.8 8e-21
Hs4758556 95.1 5e-20
SPAC29E6.02 90.5 1e-18
CE03498 89.7 2e-18
Hs22049844_1 82.4 4e-16
At1g28060 69.7 2e-12
At3g55930 54.3 1e-07
YDR473c 47.8 1e-05
At3g56790 35.8 0.037
At2g01130 29.6 2.4
7292868 29.3 3.4
At5g55670 28.5 6.1
At5g51330 28.1 7.2
7295077 27.7 9.0
SPAC1687.11 27.7 9.4
> 7293728
Length=598
Score = 97.8 bits (242), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 70/170 (41%), Positives = 97/170 (57%), Gaps = 22/170 (12%)
Query 4 QLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAP--DLARIWLSDCPAVEPWDLPLLKQT- 60
QL++ Q + ++ A KT +S KL AP D+ D PA+E WD +L Q
Sbjct 266 QLERLQNEISQIA----RKTGISSATKLALIAPKQDMP----DDVPAMEWWDSVILTQDL 317
Query 61 -----KTKAYEIDETKIDALIEHP----PPIATVAKGEAIVNMYLTPAERKKLRRRKRQE 111
+ I +T I LIEHP PP + + ++LT ERKKLRR+ R+E
Sbjct 318 ETVDDASGKISIRQTAITNLIEHPTQMKPPNEPLK--PVYLPVFLTKKERKKLRRQNRRE 375
Query 112 REREKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
+E+ +KIR+GL+ PP PK ++SNLMRVLG AV DP+K E+ VR+QMA
Sbjct 376 AWKEEQEKIRLGLVAPPEPKLRISNLMRVLGSEAVQDPTKMEQHVRDQMA 425
> Hs4758556
Length=683
Score = 95.1 bits (235), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 64/166 (38%), Positives = 92/166 (55%), Gaps = 15/166 (9%)
Query 4 QLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAPDLARIWLSDCPAVEPWDLPLLKQ---- 59
QL++ Q + ++AA KT + +L AP + D P +E WD ++
Sbjct 347 QLEKLQAEISQAA----RKTGIHTSTRLALIAPK-KELKEGDIPEIEWWDSYIIPNGFDL 401
Query 60 TKTKAYEIDETKIDALIEHP----PPIATVAKGEAIVNMYLTPAERKKLRRRKRQERERE 115
T+ D I L+EHP PP+ + +YLT E+KKLRR+ R+E ++E
Sbjct 402 TEENPKREDYFGITNLVEHPAQLNPPVDN--DTPVTLGVYLTKKEQKKLRRQTRREAQKE 459
Query 116 KPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
+K+R+GL+PPP PK ++SNLMRVLG AV DP+K E VR QMA
Sbjct 460 LQEKVRLGLMPPPEPKVRISNLMRVLGTEAVQDPTKVEAHVRAQMA 505
> SPAC29E6.02
Length=542
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 55/141 (39%), Positives = 86/141 (60%), Gaps = 11/141 (7%)
Query 28 QEKLETAAPDLARIWLSDCPAVEPWDLPLLKQTKTKAYE----ID--ETKIDALIEHPPP 81
+++L+ + + R P +E WDLP +K E ID ++ I++ I+HP P
Sbjct 224 EDELDITSKSIGR---DTIPNIEWWDLPFIKDYNDYGDENNWLIDGPQSIINSAIQHPIP 280
Query 82 IAT-VAKGE-AIVNMYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSNLMR 139
+ AK + + +++LT E+KK+RR+ R E +EK D+ +G+ PP PPK KLSNLM
Sbjct 281 VLPPYAKNQPSSHSVFLTKKEQKKIRRQTRAEARKEKQDRQLLGIEPPEPPKVKLSNLMH 340
Query 140 VLGDVAVADPSKTEKKVREQM 160
VLGD A+ DP+K E +VR+Q+
Sbjct 341 VLGDDAIKDPTKIEAEVRKQV 361
> CE03498
Length=621
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 59/166 (35%), Positives = 89/166 (53%), Gaps = 12/166 (7%)
Query 4 QLQQQQQQAAEAAAGTDTKTEVSPQEKLETAAPDLARIWLSDCPAVEPWDLPLLKQTKT- 62
+L++ Q + + AA T + V KL P + P +E WD+ +L +
Sbjct 280 KLERLQNEVSSAAQSTGISSAV----KLAMVTPTGTAKMENGVPDIEWWDMLVLDKVNYD 335
Query 63 -----KAYEIDETKIDALIEHPPPIA--TVAKGEAIVNMYLTPAERKKLRRRKRQERERE 115
E + L+EHP + T + + +YLT E+KK+RR+ R+E +E
Sbjct 336 EIPAENDMERYSQTVSELVEHPISMRAPTEPLTQQYLKVYLTTKEKKKIRRQNRKEVLKE 395
Query 116 KPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
K +KIR+GL P PK K+SNLMRVLG+ A+ DP+K E +VR+QMA
Sbjct 396 KTEKIRLGLEKAPEPKVKISNLMRVLGNEAIQDPTKMEAQVRKQMA 441
> Hs22049844_1
Length=862
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 56/159 (35%), Positives = 87/159 (54%), Gaps = 21/159 (13%)
Query 4 QLQQQQQQAAEAAAGTDTKTE-----VSPQEKLETAAPDLARIWLSDCPAVEPWDLPLLK 58
QL++ Q + ++AA TD T ++P+++L+ D P +E WD ++
Sbjct 340 QLEKLQAEISQAAQKTDIHTLTRLALIAPRKELKEG----------DIPEIEWWDSYIIP 389
Query 59 QTKTKAYEI----DETKIDALIEHPPPIATVAKGEAIVNM--YLTPAERKKLRRRKRQER 112
E D I L+EHP + + V + YLT E+KKL+R+ R+E
Sbjct 390 NGFNLREENPKREDYFGITNLVEHPAQLNAPVDNDTPVTLGVYLTKKEQKKLQRQTRREA 449
Query 113 EREKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSK 151
++E +K+R+GL+PPP PK ++SNLMRVLG AV DP+K
Sbjct 450 QKELLEKVRLGLMPPPEPKVRISNLMRVLGTEAVQDPTK 488
> At1g28060
Length=786
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/124 (39%), Positives = 65/124 (52%), Gaps = 14/124 (11%)
Query 46 CPAVEPWDLPLLKQTKTKAYEIDET----------KIDALIEHP--PPIATVAKGEAIVN 93
P VE WD +L T + EI + K+ IEHP A
Sbjct 490 IPDVEWWDANVL--TNGEYGEITDGTITESHLKIEKLTHYIEHPRPIEPPAEAAPPPPQP 547
Query 94 MYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSNLMRVLGDVAVADPSKTE 153
+ LT E+KKLR ++R +E+EK + IR GLL PP K K+SNLM+VLG A DP+K E
Sbjct 548 LKLTKKEQKKLRTQRRLAKEKEKQEMIRQGLLEPPKAKVKMSNLMKVLGSEATQDPTKLE 607
Query 154 KKVR 157
K++R
Sbjct 608 KEIR 611
> At3g55930
Length=437
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 51/92 (55%), Gaps = 2/92 (2%)
Query 72 IDALIEHPPPIATVAKGEAIVN--MYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPP 129
+ IEHP PI A+ + + +T ERKKLR +R +E EK + I G + P
Sbjct 181 FNCHIEHPFPIEPPAEAASPPPQPLKMTKEERKKLRTLRRVAKEMEKREMISQGRVEPQK 240
Query 130 PKCKLSNLMRVLGDVAVADPSKTEKKVREQMA 161
K K+SNLM+V A +P+K EK++R + A
Sbjct 241 SKVKMSNLMKVRASEATQNPTKLEKEIRTEAA 272
> YDR473c
Length=469
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 75 LIEHPPPIATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKL 134
+ HP P + + + + YLT ERK+LRR +R+ + KI++GLLP P PK KL
Sbjct 216 YVAHPLP-EKINEAKVSIKAYLTQHERKRLRRNRRKMAREAREIKIKLGLLPKPEPKVKL 274
Query 135 SNLMRVL-GDVAVADPSKTEKKVREQM 160
SN+M V D + DP+ EK V++Q+
Sbjct 275 SNMMSVFENDQNITDPTAWEKVVKDQV 301
> At3g56790
Length=206
Score = 35.8 bits (81), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 108 KRQEREREKPDKIRMGLLPPPPPKCKLSNLMRVLG 142
+R +E+EK IR L P K KLSNLM+VL
Sbjct 162 RRVAKEKEKQKMIRQALFEPQKSKVKLSNLMKVLA 196
> At2g01130
Length=1112
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 72 IDALIEHPPPIATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDKIRM 122
+ A++ PI TVA G ++ + +LTP ++K L + + R+ D + +
Sbjct 718 LGAILGCLDPILTVAAGLSVRDPFLTPQDKKDLAEAAKSQFSRDHSDHLAL 768
> 7292868
Length=787
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 16/61 (26%), Positives = 25/61 (40%), Gaps = 0/61 (0%)
Query 77 EHPPPIATVAKGEAIVNMYLTPAERKKLRRRKRQEREREKPDKIRMGLLPPPPPKCKLSN 136
E P P + V +YL +RK L++R +R+ P + PPP +
Sbjct 643 EQPAPESAETVHSVFVKVYLMDKDRKVLKKRTSLKRKDRSPIFNESMIFSVPPPSLTTTQ 702
Query 137 L 137
L
Sbjct 703 L 703
> At5g55670
Length=710
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 108 KRQEREREKPDKIRMGLLPPPPPKCKLSNLMR-VLGDV 144
+ + E + + I GLLPPPP NLMR V+G+V
Sbjct 160 RANDLEAPRGNNISQGLLPPPPVLGNNENLMRPVMGNV 197
> At5g51330
Length=635
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 35/66 (53%), Gaps = 6/66 (9%)
Query 53 DLPLLKQTKTKAY--EIDETKIDALIEHPPPIATVAKGEAIVN----MYLTPAERKKLRR 106
D+ LK+T + Y + D T+ L+E PPI T+ +VN + +P R+K R+
Sbjct 490 DMGWLKKTVDENYPKKPDSTETPLLLEDSPPIQTLEGEVKVVNKGNQITESPQNREKGRK 549
Query 107 RKRQER 112
+QER
Sbjct 550 HDQQER 555
> 7295077
Length=467
Score = 27.7 bits (60), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 5/69 (7%)
Query 13 AEAAAGTDTKTEVSPQEKLETAAPDLA--RIWLSDCPAVEPWD--LPLLKQTKTKAYEID 68
A+A +T +V QE + P +I ++D + PW P LK T + YE+D
Sbjct 141 ADAIKNFETALDVFEQEITKRGTPYFGGNKIGIADY-MIWPWFERFPALKYTLDEPYELD 199
Query 69 ETKIDALIE 77
+T+ L++
Sbjct 200 KTRYQNLLK 208
> SPAC1687.11
Length=802
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 24/34 (70%), Gaps = 2/34 (5%)
Query 96 LTPAERKKLRRRKRQEREREKPDKIR--MGLLPP 127
L+ AER KL+R +R+ +R++ + +R MG+L P
Sbjct 361 LSEAERVKLKRERRKANQRKQREIVRMQMGMLAP 394
Lambda K H
0.312 0.128 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40