bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4511_orf2
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
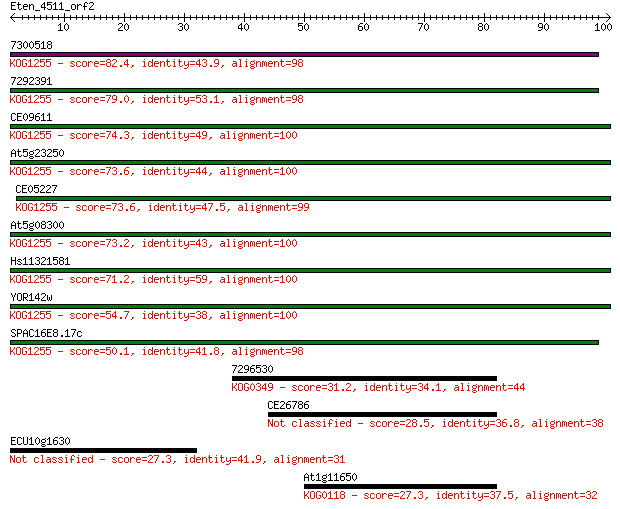
7300518 82.4 2e-16
7292391 79.0 2e-15
CE09611 74.3 5e-14
At5g23250 73.6 9e-14
CE05227 73.6 9e-14
At5g08300 73.2 1e-13
Hs11321581 71.2 5e-13
YOR142w 54.7 4e-08
SPAC16E8.17c 50.1 1e-06
7296530 31.2 0.54
CE26786 28.5 3.3
ECU10g1630 27.3 6.5
At1g11650 27.3 7.6
> 7300518
Length=342
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/98 (43%), Positives = 61/98 (62%), Gaps = 0/98 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
++FL D +GI +IGE GG++E++AA+FLK N G +AKP FI G APP ++ G
Sbjct 224 KVFLSDKEIKGIVMIGEIGGSAEEEAADFLKEKNTGCEAKPVVGFIAGQTAPPGRRMGHA 283
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFK 98
GAI GGKG K+K+ AGV + P +G+ L +
Sbjct 284 GAIISGGKGAAKDKVAALEKAGVRMTANPCHLGSTLLE 321
> 7292391
Length=324
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/98 (53%), Positives = 66/98 (67%), Gaps = 0/98 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E+FLKDP +GI LIGE GG +E+KAA++L +N G KAKP SFI G+ APP ++ G
Sbjct 219 EVFLKDPETKGIILIGEIGGVAEEKAADYLTEYNSGIKAKPVVSFIAGVSAPPGRRMGHA 278
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFK 98
GAI GGKGG +KI AGV+V P ++G LFK
Sbjct 279 GAIISGGKGGANDKIAALEKAGVIVTRSPAKMGHELFK 316
> CE09611
Length=321
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 49/100 (49%), Positives = 63/100 (63%), Gaps = 0/100 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E+FL+D +GI LIGE GG +E++AAEFLK N GS AKP SFI G+ APP ++ G
Sbjct 220 EVFLEDEQTKGIILIGEIGGQAEEQAAEFLKSRNSGSNAKPVVSFIAGVTAPPGRRMGHA 279
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKG +KI NA V+V P ++G + K
Sbjct 280 GAIIAGGKGTAGDKIEALRNANVVVTDSPAKLGVAMQKAL 319
> At5g23250
Length=341
Score = 73.6 bits (179), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 44/100 (44%), Positives = 58/100 (58%), Gaps = 3/100 (3%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E F DP EGI LIGE GG +E+ AA +K + KP +FI GL APP ++ G
Sbjct 238 EKFFVDPQTEGIVLIGEIGGTAEEDAAALIKENGTD---KPVVAFIAGLTAPPGRRMGHA 294
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKG ++KI +AGV V P ++G +F+ F
Sbjct 295 GAIVSGGKGTAQDKIKSLRDAGVKVVESPAKIGAAMFELF 334
> CE05227
Length=322
Score = 73.6 bits (179), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 47/99 (47%), Positives = 64/99 (64%), Gaps = 0/99 (0%)
Query 2 IFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSPG 61
+FL+DP +GI LIGE GG++E++AA +LK HN G+ KP SFI G+ APP ++ G G
Sbjct 220 VFLEDPETKGIILIGEIGGSAEEEAAAYLKEHNSGANRKPVVSFIAGVTAPPGRRMGHAG 279
Query 62 AIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
AI GGKG +KI AGV+V P ++GT + F
Sbjct 280 AIISGGKGTAADKINALREAGVVVTDSPAKLGTSMATAF 318
> At5g08300
Length=347
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 43/100 (43%), Positives = 58/100 (58%), Gaps = 3/100 (3%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E F DP EGI LIGE GG +E+ AA +K KP +FI GL APP ++ G
Sbjct 243 EKFFVDPQTEGIVLIGEIGGTAEEDAAALIKASG---TEKPVVAFIAGLTAPPGRRMGHA 299
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKG ++KI +AGV V P ++G+ +++ F
Sbjct 300 GAIVSGGKGTAQDKIKSLNDAGVKVVESPAKIGSAMYELF 339
> Hs11321581
Length=333
Score = 71.2 bits (173), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 59/100 (59%), Positives = 71/100 (71%), Gaps = 0/100 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
EIFL D A EGI LIGE GGN+E+ AAEFLK HN G +KP SFI GL APP ++ G
Sbjct 228 EIFLNDSATEGIILIGEIGGNAEENAAEFLKQHNSGPNSKPVVSFIAGLTAPPGRRMGHA 287
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKGG KEKI +AGV+V++ P Q+GT ++K F
Sbjct 288 GAIIAGGKGGAKEKISALQSAGVVVSMSPAQLGTTIYKEF 327
> YOR142w
Length=329
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 52/103 (50%), Gaps = 3/103 (2%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLG-SKAKPGGSFIGGLKAPPRQ--KK 57
++FL+D EGI ++GE GG +E +AA+FLK +N SK P SFI G A + +
Sbjct 223 KLFLEDETTEGIIMLGEIGGKAEIEAAQFLKEYNFSRSKPMPVASFIAGTVAGQMKGVRM 282
Query 58 GSPGAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
G GAI G + K + GV V P +G L F
Sbjct 283 GHSGAIVEGSGTDAESKKQALRDVGVAVVESPGYLGQALLDQF 325
> SPAC16E8.17c
Length=331
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/99 (41%), Positives = 57/99 (57%), Gaps = 1/99 (1%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLG-SKAKPGGSFIGGLKAPPRQKKGS 59
++FL DP +GI LIGE GG++E+ AAEF++ N S KP SFI G AP ++ G
Sbjct 226 KLFLDDPNTQGIILIGEIGGSAEEDAAEFIRAANASRSTPKPVVSFIAGATAPKGRRMGH 285
Query 60 PGAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFK 98
GAI GGKG K AGV ++ P +G+ + +
Sbjct 286 AGAIVAGGKGTAAAKFEALEAAGVRISRSPATLGSLIVE 324
> 7296530
Length=727
Score = 31.2 bits (69), Expect = 0.54, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 18/44 (40%), Gaps = 4/44 (9%)
Query 38 KAKPGGSFIGGLKAPPRQKKGSPGAIFFGGKGGGKEKIPPPPNA 81
K PG F+G +A P K +P G G P PNA
Sbjct 247 KYAPGNGFVGACQAGPEHSKANP----ITGPAAGAPSAKPAPNA 286
> CE26786
Length=598
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 2/40 (5%)
Query 44 SFIGGLKAPPRQKKGSPGAI--FFGGKGGGKEKIPPPPNA 81
SF GG + P + P + + +GGG + PPPP A
Sbjct 2 SFNGGWQPPWHNQASKPDSWRPYANNQGGGPIRQPPPPQA 41
> ECU10g1630
Length=389
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLK 31
E++ K PAP G+F G + K A EFL+
Sbjct 335 EVYKKRPAPMGVFKPGNVMSSLNKMAKEFLE 365
> At1g11650
Length=405
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 21/36 (58%), Gaps = 4/36 (11%)
Query 50 KAPPRQKKGSPGAIFFGGKGGGKEK----IPPPPNA 81
++P ++ G P ++GG G G+E+ +P PNA
Sbjct 332 RSPSNKQSGDPSQFYYGGYGQGQEQYGYTMPQDPNA 367
Lambda K H
0.315 0.145 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40