bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4540_orf2
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
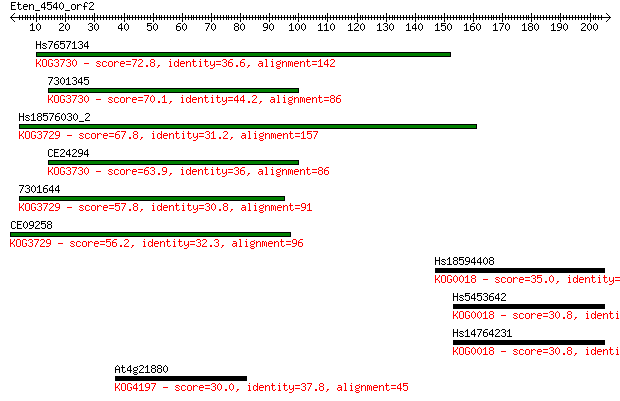
Hs7657134 72.8 4e-13
7301345 70.1 3e-12
Hs18576030_2 67.8 1e-11
CE24294 63.9 2e-10
7301644 57.8 1e-08
CE09258 56.2 5e-08
Hs18594408 35.0 0.093
Hs5453642 30.8 2.2
Hs14764231 30.8 2.2
At4g21880 30.0 3.8
> Hs7657134
Length=680
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 52/162 (32%), Positives = 75/162 (46%), Gaps = 20/162 (12%)
Query 10 FVKCVARR-YGVLEVFLEGGRSKTGLMLPPKLGLLSCAADMLFSGSVSEVLFIPISISYD 68
+VK + R Y +E FLEG RS++ L PK GLL+ + F V + +PISISYD
Sbjct 226 YVKTMLRNGYAPVEFFLEGTRSRSAKTLTPKFGLLNIVMEPFFKREVFDTYLVPISISYD 285
Query 69 RVLEAETFPQELLGEPKKAEGLSRVLKATKL-----------FAAPAYHQTL---KPSPG 114
++LE + ELLG PK E + +LKA K+ F P ++L + S
Sbjct 286 KILEETLYVYELLGVPKPKESTTGLLKARKILSENFGSIHVYFGDPVSLRSLAAGRMSRS 345
Query 115 SRGLAPQFGP-----GYRPFGTASVNVMPPMSFRDYLFSNWA 151
S L P++ P F T M + + + S W
Sbjct 346 SYNLVPRYIPQKQSEDMHAFVTEVAYKMELLQIENMVLSPWT 387
> 7301345
Length=690
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/86 (44%), Positives = 53/86 (61%), Gaps = 0/86 (0%)
Query 14 VARRYGVLEVFLEGGRSKTGLMLPPKLGLLSCAADMLFSGSVSEVLFIPISISYDRVLEA 73
VA + +E F+EG RS+ L PK+GLLS A F+G V +V+ +P+S++Y+RVLE
Sbjct 230 VANYHIGVEFFIEGTRSRNFKALVPKIGLLSMALLPYFTGEVPDVMIVPVSVAYERVLEE 289
Query 74 ETFPQELLGEPKKAEGLSRVLKATKL 99
+ F ELLG PK E KA K+
Sbjct 290 QLFVYELLGVPKPKESTKGFFKALKI 315
> Hs18576030_2
Length=383
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 49/157 (31%), Positives = 73/157 (46%), Gaps = 27/157 (17%)
Query 4 KVLLQLFVKCVARRYGVLEVFLEGGRSKTGLMLPPKLGLLSCAADMLFSGSVSEVLFIPI 63
+ LL + + R+ LE+FLEG RS++G + GLLS D L + + ++L IP+
Sbjct 68 RALLHGHIVELLRQQQFLEIFLEGTRSRSGKTSCARAGLLSVVVDTLSTNVIPDILIIPV 127
Query 64 SISYDRVLEAETFPQELLGEPKKAEGLSRVLKATKLFAAPAYHQTLKPSPGSRGLAPQFG 123
ISYDR++E + E LG+PKK E L V +RG+
Sbjct 128 GISYDRIIEGH-YNGEQLGKPKKNESLWSV---------------------ARGVIRMLR 165
Query 124 PGYRPFGTASVNVMPPMSFRDYLFSNWADEGPLKPLL 160
Y G V+ P S ++YL S + P+ LL
Sbjct 166 KNY---GCVRVDFAQPFSLKEYLESQ--SQKPVSALL 197
> CE24294
Length=513
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 50/86 (58%), Gaps = 0/86 (0%)
Query 14 VARRYGVLEVFLEGGRSKTGLMLPPKLGLLSCAADMLFSGSVSEVLFIPISISYDRVLEA 73
V +E F+E RS+ G L PK G+L + GSV +++ +P+S++YD++LE
Sbjct 217 VVNNDRTVEFFVEATRSRVGKSLHPKYGMLQMVLEPYLRGSVYDIVIVPVSMNYDKILEE 276
Query 74 ETFPQELLGEPKKAEGLSRVLKATKL 99
+ + ELLG PK E +LKA ++
Sbjct 277 KLYAYELLGFPKPKESTGALLKAREM 302
> 7301644
Length=850
Score = 57.8 bits (138), Expect = 1e-08, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 52/91 (57%), Gaps = 1/91 (1%)
Query 4 KVLLQLFVKCVARRYGVLEVFLEGGRSKTGLMLPPKLGLLSCAADMLFSGSVSEVLFIPI 63
+ L L++ ++ +E F+EGGR++TG PK G+LS + GS+ + L +P+
Sbjct 325 RAALHLYLTHALKQGHNVEFFIEGGRTRTGKPCMPKGGILSVIVNAFMDGSIPDALLVPV 384
Query 64 SISYDRVLEAETFPQELLGEPKKAEGLSRVL 94
S++Y+R+++ F +E GE K E + +
Sbjct 385 SVNYERLVDG-NFVREQKGEKKIPESFGKAI 414
> CE09258
Length=718
Score = 56.2 bits (134), Expect = 5e-08, Method: Composition-based stats.
Identities = 31/96 (32%), Positives = 53/96 (55%), Gaps = 1/96 (1%)
Query 1 KIKKVLLQLFVKCVARRYGVLEVFLEGGRSKTGLMLPPKLGLLSCAADMLFSGSVSEVLF 60
++ + +L +++ V + +E FLEG RS+ G L PK GL+S + + G + +
Sbjct 227 QLYRAILHSYIEQVLSKDMPIEFFLEGTRSRFGKALTPKNGLISNVVEAVQHGFIKDCYL 286
Query 61 IPISISYDRVLEAETFPQELLGEPKKAEGLSRVLKA 96
+P+S +YD V+E F EL+G PK E + V +
Sbjct 287 VPVSYTYDAVVEG-IFLHELMGIPKVRESVLGVFRG 321
> Hs18594408
Length=1202
Score = 35.0 bits (79), Expect = 0.093, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 27/58 (46%), Gaps = 0/58 (0%)
Query 147 FSNWADEGPLKPLLGALRHSCVSPMAQFGSLPTLTGQPQLQQQLLLLSAARRFTLWPL 204
F DE P +P L + ++CV+P +F + L+G + L LL A F P
Sbjct 1058 FQESTDENPEEPYLEGISYNCVAPGKRFMPMDNLSGGEKCVAALALLFAVHSFRPAPF 1115
> Hs5453642
Length=1233
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 153 EGPLKPLLGALRHSCVSPMAQFGSLPTLTGQPQLQQQLLLLSAARRFTLWPL 204
E P +P L + ++CV+P +F + L+G + L LL A + P
Sbjct 1101 ENPEEPYLDGINYNCVAPGKRFRPMDNLSGGEKTVAALALLFAIHSYKPAPF 1152
> Hs14764231
Length=1233
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 153 EGPLKPLLGALRHSCVSPMAQFGSLPTLTGQPQLQQQLLLLSAARRFTLWPL 204
E P +P L + ++CV+P +F + L+G + L LL A + P
Sbjct 1101 ENPEEPYLDGINYNCVAPGKRFRPMDNLSGGEKTVAALALLFAIHSYKPAPF 1152
> At4g21880
Length=859
Score = 30.0 bits (66), Expect = 3.8, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 30/49 (61%), Gaps = 4/49 (8%)
Query 37 PPKLGLLSCAADMLFSGSVSEVLFIPI-SISYDRVLE---AETFPQELL 81
P G +S A D +FS ++S+V+ +P+ SI Y+ + E + FP+ +L
Sbjct 71 PALAGQVSSALDGIFSLALSDVMIVPVKSIGYNLLRERNGEKVFPKNVL 119
Lambda K H
0.322 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3694581778
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40