bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4640_orf3
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
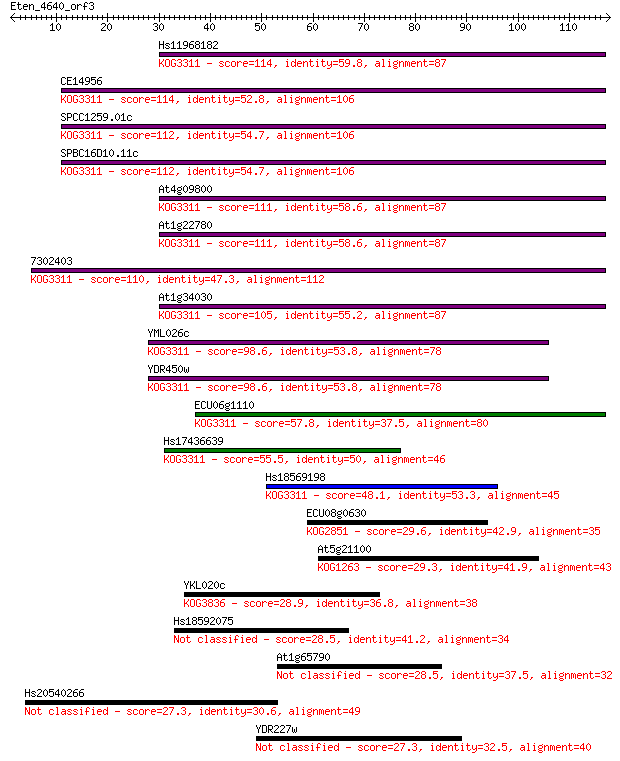
Hs11968182 114 5e-26
CE14956 114 5e-26
SPCC1259.01c 112 2e-25
SPBC16D10.11c 112 2e-25
At4g09800 111 4e-25
At1g22780 111 4e-25
7302403 110 4e-25
At1g34030 105 2e-23
YML026c 98.6 3e-21
YDR450w 98.6 3e-21
ECU06g1110 57.8 5e-09
Hs17436639 55.5 3e-08
Hs18569198 48.1 4e-06
ECU08g0630 29.6 1.5
At5g21100 29.3 1.9
YKL020c 28.9 2.6
Hs18592075 28.5 3.3
At1g65790 28.5 3.5
Hs20540266 27.3 6.7
YDR227w 27.3 7.2
> Hs11968182
Length=152
Score = 114 bits (284), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 52/87 (59%), Positives = 64/87 (73%), Gaps = 0/87 (0%)
Query 30 QIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGLRP 89
+++ I+ +P Q+ IP WFL RQKDVKDGK V AN LD+ RED ERLKK+R RGLR
Sbjct 66 RVITIMQNPRQYKIPDWFLNRQKDVKDGKYSQVLANGLDNKLREDLERLKKIRAHRGLRH 125
Query 90 YWGLKVRGRPPKTPGRPGGPVGVAKKK 116
+WGL+VRG+ KT GR G VGV+KKK
Sbjct 126 FWGLRVRGQHTKTTGRRGRTVGVSKKK 152
> CE14956
Length=154
Score = 114 bits (284), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 56/112 (50%), Positives = 73/112 (65%), Gaps = 6/112 (5%)
Query 11 AHCCCSPAA------AALLLLLLLLQIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSA 64
A CC A A L +IV I+ +P+Q+ IP+WFL RQKD+KDGK+ + +
Sbjct 41 AFVCCRKADVDVNKRAGELTEEDFDKIVTIMQNPSQYKIPNWFLNRQKDIKDGKTGQLLS 100
Query 65 NNLDSFWREDFERLKKMRLPRGLRPYWGLKVRGRPPKTPGRPGGPVGVAKKK 116
+D+ RED ER+KK+RL RGLR YWGL+VRG+ KT GR G VGV+KKK
Sbjct 101 TAVDNKLREDLERMKKIRLHRGLRHYWGLRVRGQHTKTTGRKGRTVGVSKKK 152
> SPCC1259.01c
Length=152
Score = 112 bits (279), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 58/112 (51%), Positives = 71/112 (63%), Gaps = 6/112 (5%)
Query 11 AHCCCSPAA------AALLLLLLLLQIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSA 64
A+ C A A L L +IV I+ +P+QF IPSWFL RQKD+ DGKS + A
Sbjct 41 ANIVCKKADIDMSKRAGELTTEELERIVTIIQNPSQFKIPSWFLNRQKDINDGKSFQLLA 100
Query 65 NNLDSFWREDFERLKKMRLPRGLRPYWGLKVRGRPPKTPGRPGGPVGVAKKK 116
NN+DS RED ERLKK++ RGLR L+VRG+ KT GR G VGV+KKK
Sbjct 101 NNVDSKLREDLERLKKIQTHRGLRHALDLRVRGQHTKTTGRRGKTVGVSKKK 152
> SPBC16D10.11c
Length=152
Score = 112 bits (279), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 58/112 (51%), Positives = 71/112 (63%), Gaps = 6/112 (5%)
Query 11 AHCCCSPAA------AALLLLLLLLQIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSA 64
A+ C A A L L +IV I+ +P+QF IPSWFL RQKD+ DGKS + A
Sbjct 41 ANIVCKKADIDMSKRAGELTTEELERIVTIIQNPSQFKIPSWFLNRQKDINDGKSFQLLA 100
Query 65 NNLDSFWREDFERLKKMRLPRGLRPYWGLKVRGRPPKTPGRPGGPVGVAKKK 116
NN+DS RED ERLKK++ RGLR L+VRG+ KT GR G VGV+KKK
Sbjct 101 NNVDSKLREDLERLKKIQTHRGLRHALDLRVRGQHTKTTGRRGKTVGVSKKK 152
> At4g09800
Length=152
Score = 111 bits (277), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 51/87 (58%), Positives = 61/87 (70%), Gaps = 0/87 (0%)
Query 30 QIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGLRP 89
++ IV +P QF IP WFL RQKD KDGK V +N LD R+D ERLKK+R RGLR
Sbjct 66 NLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIRNHRGLRH 125
Query 90 YWGLKVRGRPPKTPGRPGGPVGVAKKK 116
YWGL+VRG+ KT GR G VGV+KK+
Sbjct 126 YWGLRVRGQHTKTTGRRGKTVGVSKKR 152
> At1g22780
Length=152
Score = 111 bits (277), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 51/87 (58%), Positives = 61/87 (70%), Gaps = 0/87 (0%)
Query 30 QIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGLRP 89
++ IV +P QF IP WFL RQKD KDGK V +N LD R+D ERLKK+R RGLR
Sbjct 66 NLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIRNHRGLRH 125
Query 90 YWGLKVRGRPPKTPGRPGGPVGVAKKK 116
YWGL+VRG+ KT GR G VGV+KK+
Sbjct 126 YWGLRVRGQHTKTTGRRGKTVGVSKKR 152
> 7302403
Length=152
Score = 110 bits (276), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 53/112 (47%), Positives = 70/112 (62%), Gaps = 10/112 (8%)
Query 5 SLTPSAAHCCCSPAAAALLLLLLLLQIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSA 64
LT A C ++V I+ +P Q+ +P+WFL RQKD+ DGK +++
Sbjct 51 DLTKRAGECTEEEVD----------KVVTIISNPLQYKVPNWFLNRQKDIIDGKYWQLTS 100
Query 65 NNLDSFWREDFERLKKMRLPRGLRPYWGLKVRGRPPKTPGRPGGPVGVAKKK 116
+NLDS R+D ERLKK+R RGLR YWGL+VRG+ KT GR G VGV+KKK
Sbjct 101 SNLDSKLRDDLERLKKIRSHRGLRHYWGLRVRGQHTKTTGRRGRTVGVSKKK 152
> At1g34030
Length=237
Score = 105 bits (261), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 48/87 (55%), Positives = 59/87 (67%), Gaps = 0/87 (0%)
Query 30 QIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGLRP 89
++ IV +P QF IP WFL RQKD KDGK V +N LD R+D ERLKK+R RGLR
Sbjct 79 NLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIRNHRGLRH 138
Query 90 YWGLKVRGRPPKTPGRPGGPVGVAKKK 116
YWGL+VRG+ KT GR G V + +K+
Sbjct 139 YWGLRVRGQHTKTTGRRGKTVALDQKR 165
> YML026c
Length=146
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 42/78 (53%), Positives = 56/78 (71%), Gaps = 0/78 (0%)
Query 28 LLQIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGL 87
L +IV I+ +PT + IP+WFL RQ D+ DGK ANN++S R+D ERLKK+R RG+
Sbjct 66 LERIVQIMQNPTHYKIPAWFLNRQNDITDGKDYHTLANNVESKLRDDLERLKKIRAHRGI 125
Query 88 RPYWGLKVRGRPPKTPGR 105
R +WGL+VRG+ KT GR
Sbjct 126 RHFWGLRVRGQHTKTTGR 143
> YDR450w
Length=146
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 42/78 (53%), Positives = 56/78 (71%), Gaps = 0/78 (0%)
Query 28 LLQIVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGL 87
L +IV I+ +PT + IP+WFL RQ D+ DGK ANN++S R+D ERLKK+R RG+
Sbjct 66 LERIVQIMQNPTHYKIPAWFLNRQNDITDGKDYHTLANNVESKLRDDLERLKKIRAHRGI 125
Query 88 RPYWGLKVRGRPPKTPGR 105
R +WGL+VRG+ KT GR
Sbjct 126 RHFWGLRVRGQHTKTTGR 143
> ECU06g1110
Length=153
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 37 SPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGLRPYWGLKVR 96
P IP ++ Q+D+ DG + + LD+ R ER KK + R R GLKVR
Sbjct 74 DPASVGIPESYMNHQRDIIDGTTSHLIGTRLDADLRMMIERGKKNKRIRAYRLDVGLKVR 133
Query 97 GRPPKTPGRPGGPVGVAKKK 116
G+ K+ GR G +GV++KK
Sbjct 134 GQRTKSNGRRGRSMGVSRKK 153
> Hs17436639
Length=112
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 31 IVAIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANNLDSFWREDFE 76
++ ++ +P Q+ IP WFL RQKDVKDGK AN LD+ ED E
Sbjct 67 VITMMQNPHQYKIPGWFLNRQKDVKDGKYSQALANGLDNKLHEDLE 112
> Hs18569198
Length=139
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/45 (53%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 51 QKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGLRPYWGLKV 95
QKD K G V AN+LD+ RED E+LKK+ RGL +WGL+V
Sbjct 95 QKDGKAGNYSQVLANDLDNKLREDLEQLKKIPACRGLHHFWGLRV 139
> ECU08g0630
Length=298
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 59 SPPVSANNLDSFWREDFERLKK-MRLPRGLRPYWGL 93
S P+ +NLD+ EDF RLK + P L+PY +
Sbjct 255 SVPIDVSNLDTIRLEDFPRLKDVLESPERLKPYLDI 290
> At5g21100
Length=573
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 25/45 (55%), Gaps = 4/45 (8%)
Query 61 PVSANNLDSFWREDFERLKKMR-LPRGLRPYW-GLKVRGRPPKTP 103
P + N++D + E + L K LP + YW + VRGR PKTP
Sbjct 267 PFTVNDIDVYSGETYSVLLKTNALPS--KKYWISVGVRGREPKTP 309
> YKL020c
Length=1082
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 35 VGSPTQFS-IPSWFLTRQKDVKDGKSPPVSANNLDSFWR 72
+ +P FS +P F+ KD+ + S PV+ NN S R
Sbjct 474 MSTPNTFSRLPQKFIDSSKDISNHNSVPVALNNKPSIQR 512
> Hs18592075
Length=108
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 33 AIVGSPTQFSIPSWFLTRQKDVKDGKSPPVSANN 66
A+ G PTQ S+P FLTR + G P A
Sbjct 7 AVPGRPTQPSVPQSFLTRPRTQPGGHERPCHARQ 40
> At1g65790
Length=843
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 53 DVKDGKSPPVSANNLDSFWREDFERLKKMRLP 84
D++DG + + L R+ F RLK+M+LP
Sbjct 325 DLRDGSAGCMRKTRLSCDGRDGFTRLKRMKLP 356
> Hs20540266
Length=569
Score = 27.3 bits (59), Expect = 6.7, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 4 TSLTPSAAHCCCSPAAAALLLLLLLLQIVAIVGSPTQFSIPSWFLTRQK 52
TS S +C + A+ A+L ++ + IVA++ Q+ I W+L Q+
Sbjct 479 TSYCGSILYCGSTVASVAILHIVAVFYIVAVLWPLWQYFILWWYLLLQQ 527
> YDR227w
Length=1358
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 49 TRQKDVKDGKSPPVSANNLDSFWREDFERLKKMRLPRGLR 88
T K ++ KS P+ NN +SF + E++ +L LR
Sbjct 73 TSHKQLQQPKSSPLKKNNYNSFPHSNLEKISNSKLLSLLR 112
Lambda K H
0.321 0.138 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40