bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4651_orf1
Length=236
Score E
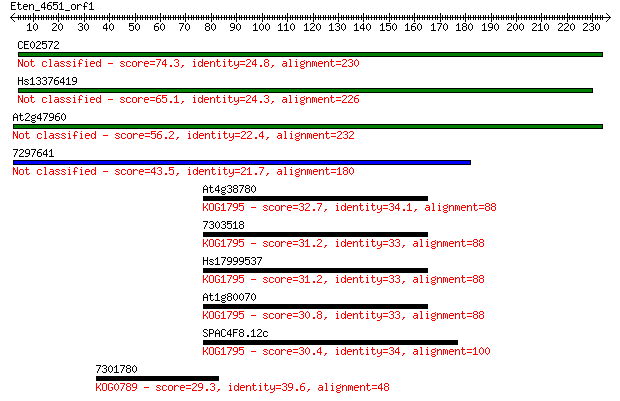
Sequences producing significant alignments: (Bits) Value
CE02572 74.3 2e-13
Hs13376419 65.1 1e-10
At2g47960 56.2 6e-08
7297641 43.5 4e-04
At4g38780 32.7 0.74
7303518 31.2 2.0
Hs17999537 31.2 2.1
At1g80070 30.8 2.2
SPAC4F8.12c 30.4 2.9
7301780 29.3 6.7
> CE02572
Length=398
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 57/231 (24%), Positives = 101/231 (43%), Gaps = 7/231 (3%)
Query 4 FKKSFRFTAQSPFKVTHRIAHLQDKTV-VECTLLNVSQQFVFLNEATILCAEGLECTRLD 62
F+K F+F P V + +++ V +E + N S +FL + + ++ T +
Sbjct 161 FRKFFKFPVSKPIDVKTKFYSAENQDVYLEAQIENTSNANMFLEKVELDPSQHYNVTSI- 219
Query 63 CCAPASESFCKGIHNFRPKDCFTVLFALFPTNPQLFLEDLAKHVPNLGQLTLQWRTSTGG 122
+ F +PKD LF L P + L K + ++G+L + WRTS G
Sbjct 220 ---AHEDEFGDVGKLLKPKDIRQFLFCLTPADVHNTLG--YKDLTSIGKLDMSWRTSMGE 274
Query 123 VGTVSDYLLTNVPQPMQPLEVRLASFPSSVEVDRPFHVELEVINRLNTALQLVLNVKLSE 182
G + L + + + + P+ V+V +PF V + N AL L L ++
Sbjct 275 KGRLQTSALQRIAPGYGDVRLSVEKTPACVDVQKPFEVSCRLYNCSERALDLQLRLEQPS 334
Query 183 LEPFVLEGPSQLTLGPLGPRSSKRFAFELLCLEPGFHSLGGIQVCDSATQQ 233
V PS ++LG L P F+ + + G S+ GI++ D+ T++
Sbjct 335 NRHLVFCSPSGVSLGQLPPSQHVDFSLNVFPVTVGIQSISGIRITDTFTKR 385
> Hs13376419
Length=354
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 55/236 (23%), Positives = 103/236 (43%), Gaps = 13/236 (5%)
Query 4 FKKSFRFTAQSPFKVTHR-------IAHLQDKTVVECTLLNVSQQFVFLNEATILCAEGL 56
F+K F+F P V + ++ + D+ +E + N++ +F+ + ++ +
Sbjct 97 FRKFFKFQVLKPLDVKTKFYNAESDLSSVTDEVFLEAQIQNMTTSPMFMEKVSLEPSIMY 156
Query 57 ECTRLDCCAPASE---SFCKGIHNFRPKDCFTVLFALFPTNPQLFLEDLAKHVPNLGQLT 113
T L+ + A E +F + +P D L+ L P N + K V +G+L
Sbjct 157 NVTELNSVSQAGECVSTFGSRAY-LQPMDTRQYLYCLKPKNEFAEKAGIIKGVTVIGKLD 215
Query 114 LQWRTSTGGVGTVSDYLLTNVPQPMQPLEVRLASFPSSVEVDRPFHVELEVINRLNTALQ 173
+ W+T+ G G + L + + + L + P +V ++ PFH+ ++ N +
Sbjct 216 IVWKTNLGERGRLQTSQLQRMAPGYGDVRLSLEAIPDTVNLEEPFHITCKITNCSERTMD 275
Query 174 LVLNVKLSELEPFVLEGPSQLTLGPLGPRSSKRFAFELLCLEPGFHSLGGIQVCDS 229
LVL ++ G S LG L P SS A LL G S+ G+++ D+
Sbjct 276 LVL--EMCNTNSIHWCGISGRQLGKLHPSSSLCLALTLLSSVQGLQSISGLRLTDT 329
> At2g47960
Length=513
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 52/253 (20%), Positives = 104/253 (41%), Gaps = 21/253 (8%)
Query 2 KSFKKSFRFTAQSPFKVTHRIAHLQDKTVVECTLLNVSQQFVFLNEATILCAEGLECTRL 61
K + F+F +P V ++ +++ T +E + N ++ +F+++ A+ RL
Sbjct 184 KYLPQFFKFVVANPLSVRTKVRVVKETTFLEACIENHTKANLFMDQVDFEPAKQWSAVRL 243
Query 62 DCCAPASESFCKGIHNFRPKDCFTVLFA------LFPTNPQLFLEDLAKHVPN--LGQLT 113
+ G+ PK + L+ NP + K + LG+
Sbjct 244 QNEDSTEDPPTSGLSGLIPKPPVIIRSGGGIHNYLYKLNPSADVSGQTKFQGSNILGKFQ 303
Query 114 LQWRTSTGGVGTVSDYLLTNVPQPMQPLEVRLASFPSSVEVDRPFHVELEVINRLNTAL- 172
+ WRT+ G G + + P + + +R+ P+ + ++RPF L + N+ + L
Sbjct 304 ITWRTNLGEPGRLQTQQILGAPVSRKEINMRVVEVPAVIHLNRPFRAYLNLTNQTDRQLG 363
Query 173 --QLVLNVKLSELE-PFVLEGPSQLTLGP-----LGPR----SSKRFAFELLCLEPGFHS 220
++ L+ ++LE P + G L L P + PR S F L+ + G
Sbjct 364 PFEVSLSQDETQLEKPVGINGLQTLELTPSKLLQMLPRIEAFGSNDFQLNLIASKLGVQK 423
Query 221 LGGIQVCDSATQQ 233
+ GI D+ ++
Sbjct 424 IAGITALDTREKK 436
> 7297641
Length=428
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 39/181 (21%), Positives = 77/181 (42%), Gaps = 4/181 (2%)
Query 2 KSFKKSFRFTAQSPFKVTHRIAHLQ-DKTVVECTLLNVSQQFVFLNEATILCAEGLECTR 60
+S +K F+F P V + + + D+ +E + NV+ L + + +E T
Sbjct 149 QSLRKFFKFQVLKPLDVKTKFYNAEIDEIYLEAQIQNVTTSPFCLEKVELDGSEDYSVTP 208
Query 61 LDCCAPASESFCKGIHNFRPKDCFTVLFALFPTNPQLFLEDLAKHVPNLGQLTLQWRTST 120
L+ P ES H +P + L+ + P D + N+G+L + WR++
Sbjct 209 LNTL-PNGESVFTVKHMLQPNNSCQFLYCIKPKGDIAKNVDTLRQFNNVGKLDIVWRSNL 267
Query 121 GGVGTVSDYLLTNVPQPMQPLEVRLASFPSSVEVDRPFHVELEVINRLNTALQLVLNVKL 180
G G + L +P + L + + +++++ F V N ++ LNV+L
Sbjct 268 GEKGRLQTSQLQRLPFECKTLRLEVLDAKNTIKIGTIFTFNCRVTNTSEHPMK--LNVRL 325
Query 181 S 181
+
Sbjct 326 A 326
> At4g38780
Length=2352
Score = 32.7 bits (73), Expect = 0.74, Method: Composition-based stats.
Identities = 30/104 (28%), Positives = 43/104 (41%), Gaps = 17/104 (16%)
Query 77 NFRPKDCFTVLFALFPTNPQLFLEDLAKHV----PNLGQLTLQWRTSTGGVGTVSDYLLT 132
N K V+F P QLFL+ + V LGQL +W+T+ V +
Sbjct 1816 NLTTKPINGVIFIFNPRTGQLFLKIIHTSVWAGQKRLGQLA-KWKTAEEVAALVRSLPVE 1874
Query 133 NVPQP--------MQPLEVRLASFPSSV----EVDRPFHVELEV 164
P+ + PLEV L FP+ V E+ PF L++
Sbjct 1875 EQPKQVIVTRKGMLDPLEVHLLDFPNIVIKGSELQLPFQACLKI 1918
> 7303518
Length=2396
Score = 31.2 bits (69), Expect = 2.0, Method: Composition-based stats.
Identities = 29/104 (27%), Positives = 42/104 (40%), Gaps = 17/104 (16%)
Query 77 NFRPKDCFTVLFALFPTNPQLFLEDLAKHV----PNLGQLTLQWRTSTGGVGTVSDYLLT 132
N K +F P QLFL+ + V LGQL +W+T+ + +
Sbjct 1857 NLTTKPINGAIFIFNPRTGQLFLKIIHTSVWAGQKRLGQLA-KWKTAEEVAALIRSLPVE 1915
Query 133 NVPQP--------MQPLEVRLASFPSSV----EVDRPFHVELEV 164
P+ + PLEV L FP+ V E+ PF L+V
Sbjct 1916 EQPKQIIVTRKGMLDPLEVHLLDFPNIVIKGSELQLPFQACLKV 1959
> Hs17999537
Length=2335
Score = 31.2 bits (69), Expect = 2.1, Method: Composition-based stats.
Identities = 29/104 (27%), Positives = 42/104 (40%), Gaps = 17/104 (16%)
Query 77 NFRPKDCFTVLFALFPTNPQLFLEDLAKHV----PNLGQLTLQWRTSTGGVGTVSDYLLT 132
N K +F P QLFL+ + V LGQL +W+T+ + +
Sbjct 1797 NLTTKPINGAIFIFNPRTGQLFLKIIHTSVWAGQKRLGQLA-KWKTAEEVAALIRSLPVE 1855
Query 133 NVPQP--------MQPLEVRLASFPSSV----EVDRPFHVELEV 164
P+ + PLEV L FP+ V E+ PF L+V
Sbjct 1856 EQPKQIIVTRKGMLDPLEVHLLDFPNIVIKGSELQLPFQACLKV 1899
> At1g80070
Length=2382
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 29/104 (27%), Positives = 42/104 (40%), Gaps = 17/104 (16%)
Query 77 NFRPKDCFTVLFALFPTNPQLFLEDLAKHV----PNLGQLTLQWRTSTGGVGTVSDYLLT 132
N K +F P QLFL+ + V LGQL +W+T+ V +
Sbjct 1844 NLTTKPINGAIFIFNPRTGQLFLKVIHTSVWAGQKRLGQLA-KWKTAEEVAALVRSLPVE 1902
Query 133 NVPQP--------MQPLEVRLASFPSSV----EVDRPFHVELEV 164
P+ + PLEV L FP+ V E+ PF L++
Sbjct 1903 EQPKQIIVTRKGMLDPLEVHLLDFPNIVIKGSELQLPFQACLKI 1946
> SPAC4F8.12c
Length=2363
Score = 30.4 bits (67), Expect = 2.9, Method: Composition-based stats.
Identities = 34/121 (28%), Positives = 50/121 (41%), Gaps = 22/121 (18%)
Query 77 NFRPKDCFTVLFALFPTNPQLFLEDLAKHV----PNLGQLTLQWRTSTGGVGTVSDYLLT 132
N K +F P QLFL+ + V LGQL +W+T+ + +
Sbjct 1821 NLTTKPINGAIFIFNPRTGQLFLKVIHTSVWAGQKRLGQLA-KWKTAEEVAALIRSLPVE 1879
Query 133 NVPQP--------MQPLEVRLASFPS----SVEVDRPFH--VELEVINRL---NTALQLV 175
P+ + PLEV L FP+ E+ PF ++L+ IN L T Q+V
Sbjct 1880 EQPRQIIVTRKGMLDPLEVHLLDFPNITIKGSELQLPFQAIIKLDKINDLILRATEPQMV 1939
Query 176 L 176
L
Sbjct 1940 L 1940
> 7301780
Length=1285
Score = 29.3 bits (64), Expect = 6.7, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 28/57 (49%), Gaps = 11/57 (19%)
Query 35 LLNVSQQFVFLNEATI---------LCAEGLECTRLDCCAPASESFCKGIHNFRPKD 82
L+ +Q++FL++A + L AE +E L C P E K I F+PKD
Sbjct 712 LVQTEEQYIFLHDALVEAIASGETNLMAEQVE--ELKNCTPYLEQQYKNIIQFQPKD 766
Lambda K H
0.323 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4669349066
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40