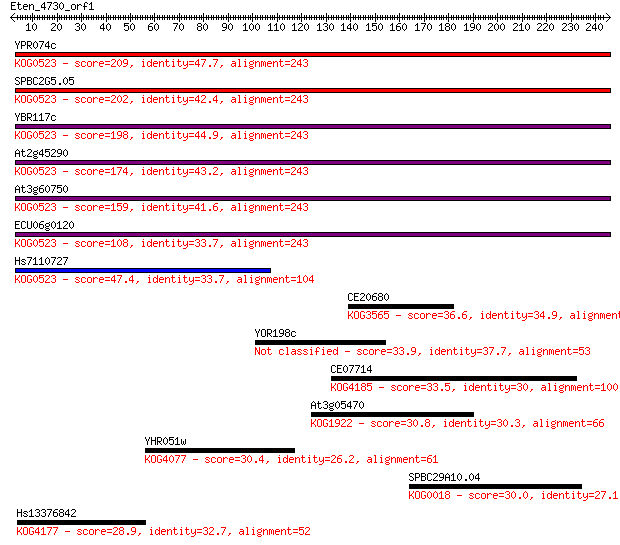
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4730_orf1
Length=245
Score E
Sequences producing significant alignments: (Bits) Value
YPR074c 209 3e-54
SPBC2G5.05 202 7e-52
YBR117c 198 7e-51
At2g45290 174 2e-43
At3g60750 159 4e-39
ECU06g0120 108 1e-23
Hs7110727 47.4 3e-05
CE20680 36.6 0.043
YOR198c 33.9 0.28
CE07714 33.5 0.44
At3g05470 30.8 2.9
YHR051w 30.4 3.1
SPBC29A10.04 30.0 4.9
Hs13376842 28.9 9.3
> YPR074c
Length=680
Score = 209 bits (533), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 116/251 (46%), Positives = 155/251 (61%), Gaps = 16/251 (6%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
HL LG LI +YD NKITIDG TS+SF E+V +R+ AYGW V++G+ D++GI +AI +
Sbjct 174 HLKLGNLIAIYDDNKITIDGATSISFDEDVAKRYEAYGWEVLYVENGNEDLAGIAKAIAQ 233
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
AK DKP+LI++ TTIG+GS + G+ VHGAPL ADD+KQLK K G NP++ F VP++
Sbjct 234 AKLSKDKPTLIKMTTTIGYGSLHAGSHSVHGAPLKADDVKQLKSKFGFNPDKSFVVPQEV 293
Query 123 RKFY------AGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKD 176
Y GV A NK W LF++Y + +P+ EL R ++ E+ L
Sbjct 294 YDHYQKTILKPGVEANNK-----WNKLFSEYQKKFPELGAELARRLSGQLPANWESKLPT 348
Query 177 MAKAAADTPASTRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGS- 235
A D+ +TR LS L + + +PELIGGSADLT SN TR K F P P GS
Sbjct 349 Y--TAKDSAVATRKLSETVLEDVYNQLPELIGGSADLTPSNLTRWKEALDFQP-PSSGSG 405
Query 236 -YADRYIHFGV 245
Y+ RYI +G+
Sbjct 406 NYSGRYIRYGI 416
> SPBC2G5.05
Length=685
Score = 202 bits (513), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 103/244 (42%), Positives = 148/244 (60%), Gaps = 3/244 (1%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
HL L LI ++D+NKITIDG TS+SF E+V +RF AYGW+ V +GD D+ GI + R
Sbjct 179 HLKLSNLIAVWDNNKITIDGATSMSFDEDVEKRFEAYGWNIVRVANGDTDLDGIEKGFRE 238
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
A + TDKP+LI +KTTIG+GS+ +GT VHG+PL +D +K+ G +P + F VP +
Sbjct 239 AMSCTDKPTLINLKTTIGYGSELQGTHSVHGSPLKPEDCVHVKKLFGFDPTKTFQVPPEV 298
Query 123 RKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAA 182
+Y A + + ++ ++ Y ++YP +L R+ R+ E E L
Sbjct 299 YAYYKERVAIASSAEEEYKKMYASYKQSYPDLSNQLERILSRKFPEGWEKHLP--VYKPG 356
Query 183 DTPASTRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFS-PNPKFGSYADRYI 241
D +TR LS L+A+ +PEL+GGSADLT SN TR +G F P+ K G+YA RYI
Sbjct 357 DKAVATRKLSEIVLDALCPVLPELVGGSADLTPSNLTRWEGAADFQPPSSKLGTYAGRYI 416
Query 242 HFGV 245
+G+
Sbjct 417 RYGI 420
> YBR117c
Length=681
Score = 198 bits (504), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 109/246 (44%), Positives = 149/246 (60%), Gaps = 6/246 (2%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
HL LG LI YDSN I+IDG TS SF E+VL+R+ AYGW V GD D+ I A+ +
Sbjct 174 HLQLGNLITFYDSNSISIDGKTSYSFDEDVLKRYEAYGWEVMEVDKGDDDMESISSALEK 233
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
AK DKP++I++ TTIG+GS +GT VHG+ L ADD+KQLK++ G +PN+ F VP++
Sbjct 234 AKLSKDKPTIIKVTTTIGFGSLQQGTAGVHGSALKADDVKQLKKRWGFDPNKSFVVPQEV 293
Query 123 RKFY-AGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAA 181
+Y V + + W +F +Y +P++ +EL R E+ E E K + K
Sbjct 294 YDYYKKTVVEPGQKLNEEWDRMFEEYKTKFPEKGKELQRRLNGELPEGWE---KHLPKFT 350
Query 182 ADTPA-STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPN-PKFGSYADR 239
D A +TR S + L + +PELIGGSADLT SN TR +G F P + G+YA R
Sbjct 351 PDDDALATRKTSQQVLTNMVQVLPELIGGSADLTPSNLTRWEGAVDFQPPITQLGNYAGR 410
Query 240 YIHFGV 245
YI +GV
Sbjct 411 YIRYGV 416
> At2g45290
Length=741
Score = 174 bits (440), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 105/245 (42%), Positives = 140/245 (57%), Gaps = 10/245 (4%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
H GLG+LI YD N I+IDGDT ++FTE+V +RF A GWH VK+G+ I AIR
Sbjct 250 HWGLGKLIAFYDDNHISIDGDTDIAFTESVDKRFEALGWHVIWVKNGNNGYDEIRAAIRE 309
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTE-KVHGAPLSADDIKQLKEKLGLNPNEDFHVPED 121
AKAVTDKP+LI++ TTIG+GS N+ VHGA L +++ + LG P E FHVPED
Sbjct 310 AKAVTDKPTLIKVTTTIGYGSPNKANSYSVHGAALGEKEVEATRNNLGW-PYEPFHVPED 368
Query 122 TRKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAA 181
+ ++ A W F Y + YP+E EL + E+ E AL
Sbjct 369 VKSHWSRHTPEGAALEADWNAKFAAYEKKYPEEAAELKSIISGELPVGWEKALPTYTP-- 426
Query 182 ADTPA-STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYADRY 240
D+P +TR LS + LNA+ +P +GGSADL +SN T LK AF N + + +R
Sbjct 427 -DSPGDATRNLSQQCLNALAKAVPGFLGGSADLASSNMTMLK---AFG-NFQKATPEERN 481
Query 241 IHFGV 245
+ FGV
Sbjct 482 LRFGV 486
> At3g60750
Length=754
Score = 159 bits (403), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 101/258 (39%), Positives = 139/258 (53%), Gaps = 23/258 (8%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
H GLG+LI YD N I+IDGDT ++FTENV QRF A GWH VK+G+ I AI+
Sbjct 250 HWGLGKLIAFYDDNHISIDGDTEIAFTENVDQRFEALGWHVIWVKNGNTGYDEIRAAIKE 309
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTE-KVHGAPLSADDIKQLKEKLGLNPNEDFHVPED 121
AK VTDKP+LI++ TTIG+GS N+ VHGA L +++ + LG P E F VP+D
Sbjct 310 AKTVTDKPTLIKVTTTIGYGSPNKANSYSVHGAALGEKEVEATRNNLGW-PYEPFQVPDD 368
Query 122 TRKF---------YAGVAARNKAE----YDAWQTLFTKYGEAYPKEKQELGRMFGREVSE 168
+ ++ +R+ E W F Y + YP+E EL + E+
Sbjct 369 VKSCLDDLLFDLHFSSHWSRHTPEGATLESDWSAKFAAYEKKYPEEASELKSIITGELPA 428
Query 169 QVETALKDMAKAAADTPA-STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAF 227
E AL ++P +TR LS + LNA+ +P +GGSADL +SN T LK F
Sbjct 429 GWEKALPTYTP---ESPGDATRNLSQQCLNALAKVVPGFLGGSADLASSNMTLLKAFGDF 485
Query 228 SPNPKFGSYADRYIHFGV 245
+ + +R + FGV
Sbjct 486 ----QKATPEERNLRFGV 499
> ECU06g0120
Length=628
Score = 108 bits (270), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 82/243 (33%), Positives = 128/243 (52%), Gaps = 23/243 (9%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
+L L ++ +YD NK TIDG TSLS E+V QRF + G+ V DGD D+ GI +A+ +
Sbjct 159 NLKLDNIVFIYDFNKTTIDGPTSLSMNEDVAQRFLSLGFEVDIV-DGD-DLDGIRKALSK 216
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
KP +I ++T IG GS EG+ K HGAPL D +++LKEK G+ P DF+V
Sbjct 217 K---VSKPMVIILETIIGRGSTVEGSCKAHGAPLGEDGVRRLKEKFGI-PAVDFYVSPSL 272
Query 123 RKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAA 182
+ + + + + W K + + ++ ++++ K K A
Sbjct 273 MEKFKILNEKKRRIRTEWDV----------KGIKAITVREWTDIQRKMDSIYKSTYK-AQ 321
Query 183 DTPASTRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYADRYIH 242
D P +TR +AL +K+ + LIGG+ADL +S T++ G+ + N G YI+
Sbjct 322 DAPKATRKHFSEALKDLKE-VSYLIGGAADLQSSVLTKIGGDDFNAENHNGG-----YIN 375
Query 243 FGV 245
FG+
Sbjct 376 FGI 378
> Hs7110727
Length=557
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 53/109 (48%), Gaps = 7/109 (6%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQR-FNAYGWHTQTVKDGDRDISGILQAIR 61
+ L L+ ++D N++ G N+ QR A+GW+T V DG RD+ + Q
Sbjct 87 YYSLDNLVAIFDVNRLGHSGALPAEHCINIYQRRCEAFGWNTYVV-DG-RDVEALCQVFW 144
Query 62 RAKAVTDKPSLIEIKTTIGWGSQN-EGTEKVHGAPLS---ADDIKQLKE 106
+A V KP+ + KT G G+ + E E H P+ AD I +L E
Sbjct 145 QASQVKHKPTAVVAKTFKGRGTPSIEDAESWHAKPMPRERADAIIKLIE 193
> CE20680
Length=1059
Score = 36.6 bits (83), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 28/43 (65%), Gaps = 1/43 (2%)
Query 139 AWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAA 181
W TL + E Y K++Q LG ++G++++ +ET +D+AK +
Sbjct 130 VWHTLVEQTKEEY-KKRQSLGELYGKQMTASIETRCEDLAKIS 171
> YOR198c
Length=470
Score = 33.9 bits (76), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 101 IKQLKEKL-GLNPNEDFHVPEDTRKFYAGVAARNKAEYDAWQTLFTKYGEAYPK 153
I QLKE+L GLNP + + E+ ++ + ++ + YD QTLF K Y K
Sbjct 167 INQLKEELNGLNPKDVSNQFEENQQKLNDIHSKTQGVYDKRQTLFNKRAALYKK 220
> CE07714
Length=331
Score = 33.5 bits (75), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 48/119 (40%), Gaps = 20/119 (16%)
Query 132 RNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAADTPASTRVL 191
+N E QTLF + GE + K+K ++ + + + E+V K+ D + RV
Sbjct 189 QNVEELKKAQTLFNENGELFQKKKNQITNFYSK-LREKVNEREKEALSELKDVARTARVN 247
Query 192 SGKA---LNAIKDFMPELIGGS----------------ADLTTSNETRLKGETAFSPNP 231
+ K+ L ++ ELI + A L SN + E F PNP
Sbjct 248 NTKSLLTLMQVRSIYDELISEAKELLKKTSATSFREVEAILNRSNAHIMSQEKIFEPNP 306
> At3g05470
Length=884
Score = 30.8 bits (68), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 35/68 (51%), Gaps = 7/68 (10%)
Query 124 KFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAAD 183
+F+ VA N E D W+ L + GE Y + +GR G E E+++ + + +A D
Sbjct 22 RFFCIVA--NAKELDDWKVLTVENGERY---RTHVGRYAGEEGGEKIKLRVLEKFRALLD 76
Query 184 T--PASTR 189
P+++R
Sbjct 77 LIKPSTSR 84
> YHR051w
Length=148
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 33/61 (54%), Gaps = 5/61 (8%)
Query 56 ILQAIRRAKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNED 115
I +A+R A+ V D P+ I + + + +NE K + D++K ++++LG+ E+
Sbjct 88 IEKALRAARRVNDLPTAIRVFEALKYKVENEDQYKAY-----LDELKDVRQELGVPLKEE 142
Query 116 F 116
Sbjct 143 L 143
> SPBC29A10.04
Length=1233
Score = 30.0 bits (66), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 38/71 (53%), Gaps = 1/71 (1%)
Query 164 REVSEQVETALKDMAKAAADTPASTR-VLSGKALNAIKDFMPELIGGSADLTTSNETRLK 222
+E++E++++ L+ + +A+AD S + +AL A+K PE+ G DL T + + +
Sbjct 479 QELNEELQSCLQKILEASADRNESKQDAKKREALYALKRIYPEVKGRIIDLCTPTQKKYE 538
Query 223 GETAFSPNPKF 233
A + F
Sbjct 539 SAIAAALGKNF 549
> Hs13376842
Length=1166
Score = 28.9 bits (63), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 4 LGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISG 55
LGL L+ +++ +IT+D + E NAYG + +K +R ISG
Sbjct 885 LGLEHLMDIFEREQITLDVLVEMGHKELKEIGINAYGHRHKLIKGVERLISG 936
Lambda K H
0.314 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4938804274
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40