bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4738_orf1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
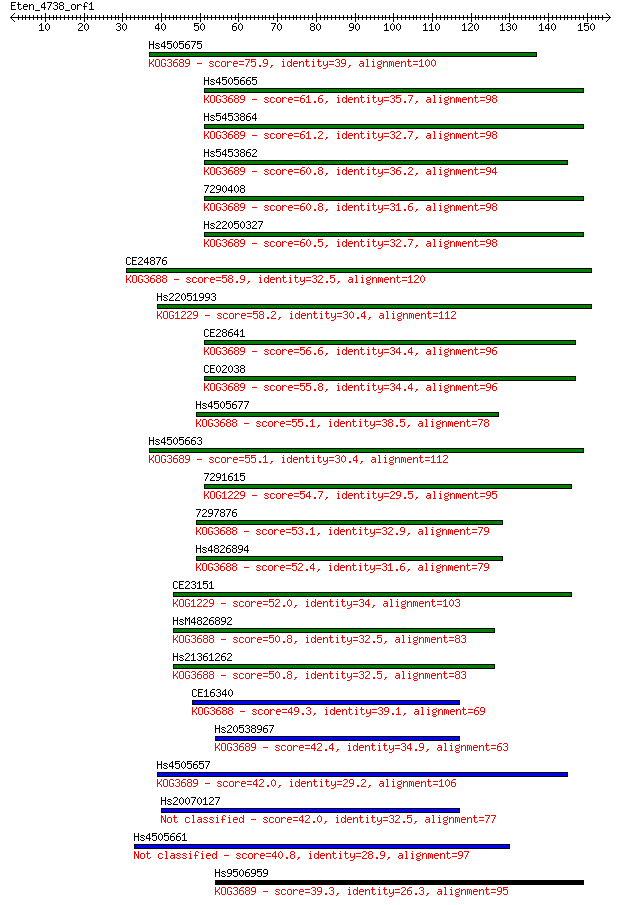
Hs4505675 75.9 3e-14
Hs4505665 61.6 7e-10
Hs5453864 61.2 8e-10
Hs5453862 60.8 1e-09
7290408 60.8 1e-09
Hs22050327 60.5 1e-09
CE24876 58.9 4e-09
Hs22051993 58.2 7e-09
CE28641 56.6 2e-08
CE02038 55.8 3e-08
Hs4505677 55.1 5e-08
Hs4505663 55.1 5e-08
7291615 54.7 6e-08
7297876 53.1 2e-07
Hs4826894 52.4 3e-07
CE23151 52.0 4e-07
HsM4826892 50.8 1e-06
Hs21361262 50.8 1e-06
CE16340 49.3 3e-06
Hs20538967 42.4 3e-04
Hs4505657 42.0 4e-04
Hs20070127 42.0 5e-04
Hs4505661 40.8 0.001
Hs9506959 39.3 0.003
> Hs4505675
Length=593
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 39/101 (38%), Positives = 61/101 (60%), Gaps = 2/101 (1%)
Query 37 FLNSDEDH-TLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVF 95
F S+E+H TLL LI C DISN + P W +++E+ Q + E+ +GLPV F
Sbjct 442 FDYSNEEHMTLLKMILIKCCDISNEVRPMEVAEPWVDCLLEEYFMQSDREKSEGLPVAPF 501
Query 96 MDARTELLRTQSQIGFLSFVVLDQFRALSDLVPGAEELVVQ 136
MD R ++ + +QIGF+ FV++ F ++ L P EE+++Q
Sbjct 502 MD-RDKVTKATAQIGFIKFVLIPMFETVTKLFPMVEEIMLQ 541
> Hs4505665
Length=712
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 55/99 (55%), Gaps = 2/99 (2%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
L+HCAD+SNP P QW I+ EF Q + ER GL ++ D T + +SQ+G
Sbjct 541 LVHCADLSNPTKPLPLYRQWTDRIMAEFFQQGDRERESGLDISPMCDKHTASVE-KSQVG 599
Query 111 FLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDWQAAM 148
F+ ++ + +DLV P A++L+ E N E +Q+ +
Sbjct 600 FIDYIAHPLWETWADLVHPDAQDLLDTLEDNREWYQSKI 638
> Hs5453864
Length=809
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 58/99 (58%), Gaps = 2/99 (2%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
++HCAD+SNP P + QW I++EF Q + ER +G+ ++ D + +SQ+G
Sbjct 615 MVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGMEISPMCDKHNASVE-KSQVG 673
Query 111 FLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDWQAAM 148
F+ ++V + +DLV P A++++ E N E +Q+ +
Sbjct 674 FIDYIVHPLWETWADLVHPDAQDILDTLEDNREWYQSTI 712
> Hs5453862
Length=647
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 54/95 (56%), Gaps = 3/95 (3%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
++HCAD+SNP P QW I+ EF Q + ER +G+ ++ D T + +SQ+G
Sbjct 347 MVHCADLSNPTKPLELYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTASVE-KSQVG 405
Query 111 FLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDW 144
F+ ++V + +DLV P A+E++ E N DW
Sbjct 406 FIDYIVHPLWETWADLVHPDAQEILDTLEDN-RDW 439
> 7290408
Length=624
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 59/99 (59%), Gaps = 2/99 (2%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
L+HCAD+SNP P +W +L+++EF Q + ER G+ ++ D + +SQ+G
Sbjct 439 LVHCADLSNPTKPLPLYKRWVALLMEEFFLQGDKERESGMDISPMCDRHNATIE-KSQVG 497
Query 111 FLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDWQAAM 148
F+ ++V + +DLV P A++++ E+N + +Q+ +
Sbjct 498 FIDYIVHPLWETWADLVHPDAQDILDTLEENRDYYQSMI 536
> Hs22050327
Length=641
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 58/99 (58%), Gaps = 2/99 (2%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
++HCAD+SNP P + QW I++EF Q + ER +G+ ++ D + +SQ+G
Sbjct 447 MVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGMEISPMCDKHNASVE-KSQVG 505
Query 111 FLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDWQAAM 148
F+ ++V + +DLV P A++++ E N E +Q+ +
Sbjct 506 FIDYIVHPLWETWADLVHPDAQDILDTLEDNREWYQSTI 544
> CE24876
Length=562
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 39/127 (30%), Positives = 66/127 (51%), Gaps = 8/127 (6%)
Query 31 ERTTGAFLNSDEDHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGL 90
ER T + + D L+ LI ADI++P P + QW I +EF Q + ER +GL
Sbjct 285 ERLTEIDVQVETDRLLIGKLLIKMADINSPTKPYGLHRQWTDRICEEFYEQGDDERRRGL 344
Query 91 PVTVFMDARTELLRTQSQIGFLSFVVLDQFRALSD-----LVPGAE--ELVVQGEKNLED 143
P+T +MD R + + Q F++ VV A+++ ++PG + EL++ E N
Sbjct 345 PITPYMD-RGDAQVAKLQDSFIAHVVSPLATAMNECGLLPILPGLDTSELIINMEHNHRK 403
Query 144 WQAAMDI 150
W+ +++
Sbjct 404 WKEQIEL 410
> Hs22051993
Length=761
Score = 58.2 bits (139), Expect = 7e-09, Method: Composition-based stats.
Identities = 34/112 (30%), Positives = 56/112 (50%), Gaps = 1/112 (0%)
Query 39 NSDEDHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDA 98
N E+ L+ +I CAD++NP P ++WA I +E+ AQ + E+ +GLPV + +
Sbjct 640 NFPENQILIKRMMIKCADVANPCRPLDLCIEWAGRISEEYFAQTDEEKRQGLPVVMPVFD 699
Query 99 RTELLRTQSQIGFLSFVVLDQFRALSDLVPGAEELVVQGEKNLEDWQAAMDI 150
R +SQI F+ + + D F A D L+ N + W+ D+
Sbjct 700 RNTCSIPKSQISFIDYFITDMFDAW-DAFAHLPALMQHLADNYKHWKTLDDL 750
> CE28641
Length=626
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 56/97 (57%), Gaps = 2/97 (2%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
+IH AD+SNP P QW I++E+ Q + E+ GL ++ D R + +SQ+G
Sbjct 512 MIHLADLSNPTKPIELYQQWNQRIMEEYWRQGDKEKELGLEISPMCD-RGNVTIEKSQVG 570
Query 111 FLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDWQA 146
F+ ++V + +DLV P A+ ++ Q E+N E +Q+
Sbjct 571 FIDYIVHPLYETWADLVYPDAQNILDQLEENREWYQS 607
> CE02038
Length=549
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 56/97 (57%), Gaps = 2/97 (2%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
+IH AD+SNP P QW I++E+ Q + E+ GL ++ D R + +SQ+G
Sbjct 435 MIHLADLSNPTKPIELYQQWNQRIMEEYWRQGDKEKELGLEISPMCD-RGNVTIEKSQVG 493
Query 111 FLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDWQA 146
F+ ++V + +DLV P A+ ++ Q E+N E +Q+
Sbjct 494 FIDYIVHPLYETWADLVYPDAQNILDQLEENREWYQS 530
> Hs4505677
Length=536
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 44/78 (56%), Gaps = 1/78 (1%)
Query 49 TCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQ 108
+ L+H ADIS+P + +W +++EF Q + E GLP + D RT L QSQ
Sbjct 363 SLLLHAADISHPTKQWLVHSRWTKALMEEFFRQGDKEAELGLPFSPLCD-RTSTLVAQSQ 421
Query 109 IGFLSFVVLDQFRALSDL 126
IGF+ F+V F L+D+
Sbjct 422 IGFIDFIVEPTFSVLTDV 439
> Hs4505663
Length=564
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 34/113 (30%), Positives = 61/113 (53%), Gaps = 3/113 (2%)
Query 37 FLNSDEDHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFM 96
L++ D + ++HCAD+SNP QW I++EF Q + ER +G+ ++
Sbjct 373 LLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEISPMC 432
Query 97 DARTELLRTQSQIGFLSFVVLDQFRALSDLV-PGAEELVVQGEKNLEDWQAAM 148
D T + +SQ+GF+ ++V + +DLV P A++++ E N +W +M
Sbjct 433 DKHTASVE-KSQVGFIDYIVHPLWETWADLVQPDAQDILDTLEDN-RNWYQSM 483
> 7291615
Length=1060
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 49/95 (51%), Gaps = 1/95 (1%)
Query 51 LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIG 110
LI AD+SNP P + ++WA I +E+ Q + E+ + LP+ + M R +SQIG
Sbjct 940 LIKVADVSNPARPMQFCIEWARRIAEEYFMQTDEEKQRHLPIVMPMFDRATCSIPKSQIG 999
Query 111 FLSFVVLDQFRALSDLVPGAEELVVQGEKNLEDWQ 145
F+ +++ D A + +L+ + N W+
Sbjct 1000 FIEYIIQDMMHAWESFI-DMPQLITYMQINYSQWK 1033
> 7297876
Length=605
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 45/79 (56%), Gaps = 1/79 (1%)
Query 49 TCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQ 108
+ ++HC DIS+P + +W L+++EF Q ++E+ GLP + D R L +SQ
Sbjct 381 SLVLHCCDISHPAKQWGVHHRWTMLLLEEFFRQGDLEKELGLPFSPLCD-RNNTLVAESQ 439
Query 109 IGFLSFVVLDQFRALSDLV 127
I F+ F+V +SD++
Sbjct 440 ICFIDFIVEPSMGVMSDML 458
> Hs4826894
Length=634
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 45/79 (56%), Gaps = 1/79 (1%)
Query 49 TCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQ 108
+ ++H ADIS+P + +W +++EF Q + E GLP + D ++ ++ QSQ
Sbjct 369 SLMLHTADISHPAKAWDLHHRWTMSLLEEFFRQGDREAELGLPFSPLCDRKSTMV-AQSQ 427
Query 109 IGFLSFVVLDQFRALSDLV 127
+GF+ F+V F L+D+
Sbjct 428 VGFIDFIVEPTFTVLTDMT 446
> CE23151
Length=760
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 49/105 (46%), Gaps = 3/105 (2%)
Query 43 DHTLLATC--LIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDART 100
D L C L+ CADISNP +WA I++E+ Q E+ KGLPVT+ + R
Sbjct 641 DTNSLTICDMLVKCADISNPAREWGLCQRWAHRIVEEYFEQTREEKEKGLPVTMEVFDRN 700
Query 101 ELLRTQSQIGFLSFVVLDQFRALSDLVPGAEELVVQGEKNLEDWQ 145
+Q GF+ + F ++ EL Q E N E W+
Sbjct 701 TCNVPITQCGFIDMFAREAFATFTEFAKLG-ELSDQLESNYEKWK 744
> HsM4826892
Length=535
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 46/83 (55%), Gaps = 1/83 (1%)
Query 43 DHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTEL 102
D + ++H ADIS+P + + +W +++EF Q + E GLP + D ++ +
Sbjct 353 DRAKTMSLILHAADISHPAKSWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRKSTM 412
Query 103 LRTQSQIGFLSFVVLDQFRALSD 125
+ QSQIGF+ F+V F L+D
Sbjct 413 V-AQSQIGFIDFIVEPTFSLLTD 434
> Hs21361262
Length=545
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 46/83 (55%), Gaps = 1/83 (1%)
Query 43 DHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTEL 102
D + ++H ADIS+P + + +W +++EF Q + E GLP + D ++ +
Sbjct 353 DRAKTMSLILHAADISHPAKSWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRKSTM 412
Query 103 LRTQSQIGFLSFVVLDQFRALSD 125
+ QSQIGF+ F+V F L+D
Sbjct 413 V-AQSQIGFIDFIVEPTFSLLTD 434
> CE16340
Length=664
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query 48 ATCLI-HCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQ 106
A CLI H DIS+P P + +W +++EF Q ++E GLP + D T +
Sbjct 471 ALCLIVHACDISHPAKPWNLHERWTEGVLEEFFRQGDLEASMGLPYSPLCDRHT-VHVAD 529
Query 107 SQIGFLSFVV 116
SQIGF+ F+V
Sbjct 530 SQIGFIDFIV 539
> Hs20538967
Length=418
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 54 CADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIGFLS 113
CADI NP + QW+ + +EF Q ++E+ L V+ D TE + QIGF++
Sbjct 296 CADICNPCRTWELSKQWSEKVTEEFFHQGDIEKKYHLGVSPLCDRHTESI-ANIQIGFMT 354
Query 114 FVV 116
++V
Sbjct 355 YLV 357
> Hs4505657
Length=941
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 51/106 (48%), Gaps = 1/106 (0%)
Query 39 NSDEDHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDA 98
N+ + H LL L+ D+S+ + + A LI +EF +Q ++E+ G MD
Sbjct 791 NNKQHHRLLLCLLMTSCDLSDQTKGWKTTRKIAELIYKEFFSQGDLEKAMGNRPMEMMD- 849
Query 99 RTELLRTQSQIGFLSFVVLDQFRALSDLVPGAEELVVQGEKNLEDW 144
R + + QI F+ + + ++ L DL P A EL + N E W
Sbjct 850 REKAYIPELQISFMEHIAMPIYKLLQDLFPKAAELYERVASNREHW 895
> Hs20070127
Length=1141
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 1/77 (1%)
Query 40 SDEDHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDAR 99
++ D L+ I ADI+ P + ++QW I+ EF Q + E GLP++ FMD R
Sbjct 934 NENDRLLVCQMCIKLADINGPAKCKELHLQWTDGIVNEFYEQGDEEASLGLPISPFMD-R 992
Query 100 TELLRTQSQIGFLSFVV 116
+ Q F+S +V
Sbjct 993 SAPQLANLQESFISHIV 1009
> Hs4505661
Length=1112
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 5/100 (5%)
Query 33 TTGAFLNSDEDHTLLATCLIHCADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPV 92
+ G +++ D L+ I ADI+ P +++W I+ EF Q + E + GLP+
Sbjct 914 SNGIEWSNENDRLLVCQVCIKLADINGPAKVRDLHLKWTEGIVNEFYEQGDEEANLGLPI 973
Query 93 TVFMDARTELLRTQSQIGFLSFVV---LDQFRALSDLVPG 129
+ FMD R+ + Q F++ +V + + A + L+PG
Sbjct 974 SPFMD-RSSPQLAKLQESFITHIVGPLCNSYDA-AGLLPG 1011
> Hs9506959
Length=450
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query 54 CADISNPLLPERRNMQWASLIIQEFNAQVEMERHKGLPVTVFMDARTELLRTQSQIGFLS 113
CADI NP + QW+ + +EF Q E+E+ L ++ + + + + + QIGF+S
Sbjct 321 CADICNPCRIWEMSKQWSERVCEEFYRQGELEQKFELEISPLCNQQKDSIPS-IQIGFMS 379
Query 114 FVVLDQFRALSDLVPG---AEELVVQGEKNLEDWQAAM 148
++V FR + +E ++ N W++ +
Sbjct 380 YIVEPLFREWAHFTGNSTLSENMLGHLAHNKAQWKSLL 417
Lambda K H
0.319 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40