bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4872_orf1
Length=81
Score E
Sequences producing significant alignments: (Bits) Value
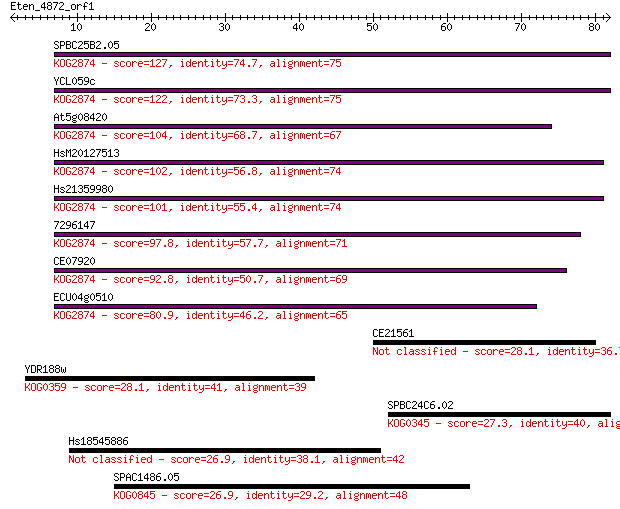
SPBC25B2.05 127 4e-30
YCL059c 122 2e-28
At5g08420 104 5e-23
HsM20127513 102 1e-22
Hs21359980 101 3e-22
7296147 97.8 4e-21
CE07920 92.8 1e-19
ECU04g0510 80.9 5e-16
CE21561 28.1 3.8
YDR188w 28.1 4.3
SPBC24C6.02 27.3 6.5
Hs18545886 26.9 9.9
SPAC1486.05 26.9 10.0
> SPBC25B2.05
Length=327
Score = 127 bits (320), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 56/75 (74%), Positives = 65/75 (86%), Gaps = 0/75 (0%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNVHPVYHVKELLIRRELAKDEKLKNENWD 66
CYILVQG TVA MG KGLK+V R+VEDCM N+HP+YH+KEL+I+RELAKD L NE+WD
Sbjct 175 CYILVQGTTVAVMGGYKGLKEVRRIVEDCMHNIHPIYHIKELMIKRELAKDPTLANESWD 234
Query 67 RFLPQFKKRNVQRKK 81
RFLPQFKKRNV R+K
Sbjct 235 RFLPQFKKRNVARRK 249
> YCL059c
Length=316
Score = 122 bits (305), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 55/75 (73%), Positives = 65/75 (86%), Gaps = 0/75 (0%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNVHPVYHVKELLIRRELAKDEKLKNENWD 66
CYILVQG TV+AMG KGLK+V RVVEDCM+N+HP+YH+KEL+I+RELAK +L NE+W
Sbjct 162 CYILVQGNTVSAMGPFKGLKEVRRVVEDCMKNIHPIYHIKELMIKRELAKRPELANEDWS 221
Query 67 RFLPQFKKRNVQRKK 81
RFLP FKKRNV RKK
Sbjct 222 RFLPMFKKRNVARKK 236
> At5g08420
Length=391
Score = 104 bits (259), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 46/68 (67%), Positives = 57/68 (83%), Gaps = 1/68 (1%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNV-HPVYHVKELLIRRELAKDEKLKNENW 65
CYILVQG TVAAMG KGLKQ+ R+VEDC+QN+ HPVYH+K L++++EL KD L NE+W
Sbjct 180 CYILVQGSTVAAMGPFKGLKQLRRIVEDCVQNIMHPVYHIKTLMMKKELEKDPALANESW 239
Query 66 DRFLPQFK 73
DRFLP F+
Sbjct 240 DRFLPTFR 247
> HsM20127513
Length=381
Score = 102 bits (255), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 42/74 (56%), Positives = 63/74 (85%), Gaps = 0/74 (0%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNVHPVYHVKELLIRRELAKDEKLKNENWD 66
CYI+VQG TV+A+G GLK+V +VV D M+N+HP+Y++K L+I+RELAKD +L++++W+
Sbjct 181 CYIMVQGNTVSAIGPFSGLKEVRKVVLDTMKNIHPIYNIKSLMIKRELAKDSELRSQSWE 240
Query 67 RFLPQFKKRNVQRK 80
RFLPQFK +NV ++
Sbjct 241 RFLPQFKHKNVNKR 254
> Hs21359980
Length=381
Score = 101 bits (252), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 41/74 (55%), Positives = 62/74 (83%), Gaps = 0/74 (0%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNVHPVYHVKELLIRRELAKDEKLKNENWD 66
CYI+VQG TV+A+G GLK+V +V D M+N+HP+Y++K L+I+RELAKD +L++++W+
Sbjct 181 CYIMVQGNTVSAIGPFSGLKEVRKVALDTMKNIHPIYNIKSLMIKRELAKDSELRSQSWE 240
Query 67 RFLPQFKKRNVQRK 80
RFLPQFK +NV ++
Sbjct 241 RFLPQFKHKNVNKR 254
> 7296147
Length=345
Score = 97.8 bits (242), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 41/71 (57%), Positives = 57/71 (80%), Gaps = 0/71 (0%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNVHPVYHVKELLIRRELAKDEKLKNENWD 66
CY+LVQG TV+A+G KGL+QV +V + M NVHP+Y++K L+I+REL KD +L NE+W
Sbjct 163 CYVLVQGNTVSALGPYKGLQQVRDIVLETMNNVHPIYNIKALMIKRELMKDPRLANEDWS 222
Query 67 RFLPQFKKRNV 77
RFLP+FK +N+
Sbjct 223 RFLPKFKNKNI 233
> CE07920
Length=370
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 57/69 (82%), Gaps = 0/69 (0%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNVHPVYHVKELLIRRELAKDEKLKNENWD 66
CY+ VQG TV A+G GLKQ+ ++V DCM+N+HP+Y++K ++I+REL+K+++LK+ NWD
Sbjct 186 CYVCVQGGTVCAVGPLAGLKQINQIVTDCMKNIHPIYNIKTMMIKRELSKNDELKDANWD 245
Query 67 RFLPQFKKR 75
+LP ++K+
Sbjct 246 AYLPNYRKK 254
> ECU04g0510
Length=283
Score = 80.9 bits (198), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 49/65 (75%), Gaps = 0/65 (0%)
Query 7 CYILVQGQTVAAMGSPKGLKQVPRVVEDCMQNVHPVYHVKELLIRRELAKDEKLKNENWD 66
CY+LV G+TV+ +G +G+++ ++V DCM N+HP+Y +K L+ +R++ DE +NE+W
Sbjct 143 CYVLVHGKTVSIIGGFRGVEEAKKIVVDCMNNIHPMYQIKRLVEKRKMESDETKENEDWS 202
Query 67 RFLPQ 71
RFLPQ
Sbjct 203 RFLPQ 207
> CE21561
Length=308
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 50 IRRELAKDEKLKNENWDRFLPQFKKRNVQR 79
I+ +L +DE+++N NW+ FKK R
Sbjct 178 IQLKLVRDEEIRNSNWNLLETSFKKMKNSR 207
> YDR188w
Length=546
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 7/44 (15%)
Query 3 NPKPCYILVQGQTVAAMGSPK-----GLKQVPRVVEDCMQNVHP 41
+PK C IL++G T A+ K GL+ V V++D +N+ P
Sbjct 372 DPKSCTILIKGSTHYALAQTKDAVRDGLRAVANVLKD--KNIIP 413
> SPBC24C6.02
Length=606
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 52 RELAKDEKLKNENWDRFLPQFKKRNVQRKK 81
++LA+ K+KNE W + +KRN +R+K
Sbjct 506 KKLARPAKIKNEAWSKQKEVKEKRNTRREK 535
> Hs18545886
Length=960
Score = 26.9 bits (58), Expect = 9.9, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 7/48 (14%)
Query 9 ILVQGQTVAAMGSPKGLKQVPRVVEDCMQN------VHPVYHVKELLI 50
+ V G ++ SPKGL+ VPR ++ C+ + H +HV+ LI
Sbjct 429 VAVPGPDPCSLSSPKGLETVPR-LKTCLNSGPTFSRCHKPFHVQPALI 475
> SPAC1486.05
Length=1778
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Query 15 TVAAMGSPKGLKQVPRVVEDCMQNVH---PVYHVKELLIRRELAKDEKLKN 62
+ +A G PK L + +D + + P ++K+L+I +++KD+ L+N
Sbjct 707 STSASGKPKSLHLFDSLNDDVLLSADAFTPRQNIKKLVITHKISKDDILQN 757
Lambda K H
0.320 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1162440328
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40