bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4914_orf1
Length=142
Score E
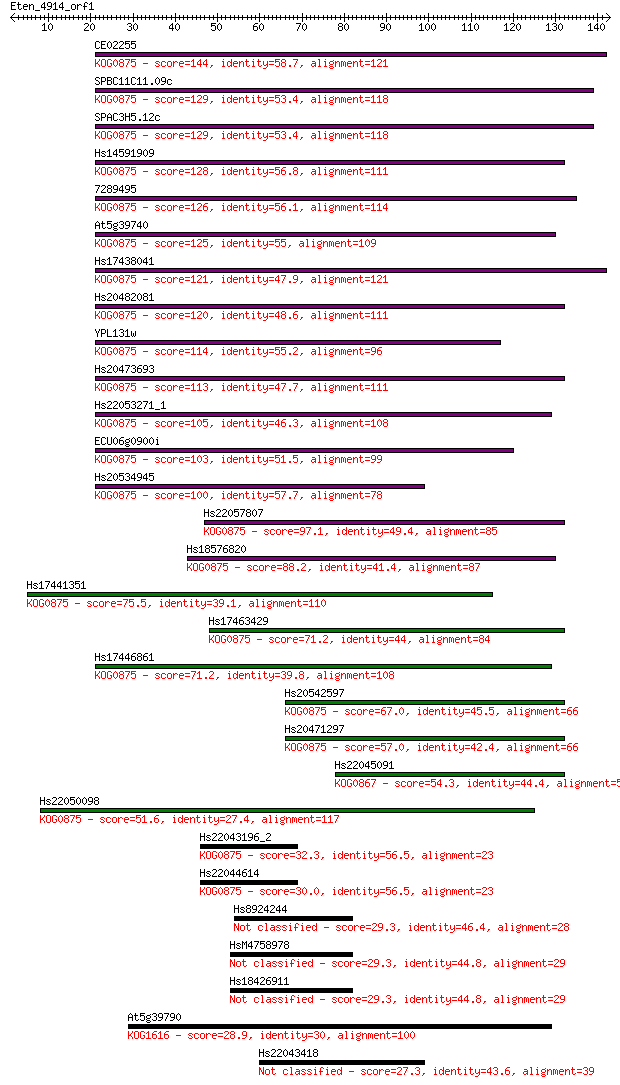
Sequences producing significant alignments: (Bits) Value
CE02255 144 6e-35
SPBC11C11.09c 129 2e-30
SPAC3H5.12c 129 2e-30
Hs14591909 128 4e-30
7289495 126 2e-29
At5g39740 125 4e-29
Hs17438041 121 5e-28
Hs20482081 120 8e-28
YPL131w 114 4e-26
Hs20473693 113 1e-25
Hs22053271_1 105 3e-23
ECU06g0900i 103 2e-22
Hs20534945 100 9e-22
Hs22057807 97.1 1e-20
Hs18576820 88.2 5e-18
Hs17441351 75.5 3e-14
Hs17463429 71.2 5e-13
Hs17446861 71.2 5e-13
Hs20542597 67.0 1e-11
Hs20471297 57.0 1e-08
Hs22045091 54.3 7e-08
Hs22050098 51.6 5e-07
Hs22043196_2 32.3 0.33
Hs22044614 30.0 1.3
Hs8924244 29.3 2.7
HsM4758978 29.3 2.8
Hs18426911 29.3 2.8
At5g39790 28.9 3.5
Hs22043418 27.3 8.9
> CE02255
Length=293
Score = 144 bits (363), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 71/121 (58%), Positives = 89/121 (73%), Gaps = 2/121 (1%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY AR+RL +QDKNKYN PK R +VR+TN V+ Q+ Y+ ++GD +V SA S EL
Sbjct 25 EGKTDYYARKRLTVQDKNKYNTPKYRLIVRITNKDVVAQLAYSKIEGDVVVASAYSHELP 84
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEVLRGSAGPSN 140
RYG+KVGLTNY+AAYATGL LARR LK GL +KG E+ +GE+Y++EE G P
Sbjct 85 RYGLKVGLTNYAAAYATGLLLARRHLKTIGLDSTYKGHEELTGEDYNVEE--EGDRAPFK 142
Query 141 A 141
A
Sbjct 143 A 143
> SPBC11C11.09c
Length=294
Score = 129 bits (323), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 63/118 (53%), Positives = 83/118 (70%), Gaps = 1/118 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY AR+RLI Q KNKYNAPK R VVR +N V CQ++ + + GD ++ A S EL
Sbjct 25 EGKTDYYARKRLIAQAKNKYNAPKYRLVVRFSNRFVTCQIVSSRVNGDYVLAHAHSSELP 84
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEVLRGSAGP 138
RYGIK GL N++AAYATGL +ARR L + GLAD+++G+ +P G E+ + E + P
Sbjct 85 RYGIKWGLANWTAAYATGLLVARRALAKVGLADKYEGVTEPEG-EFELTEAIEDGPRP 141
> SPAC3H5.12c
Length=294
Score = 129 bits (323), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 63/118 (53%), Positives = 83/118 (70%), Gaps = 1/118 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY AR+RLI Q KNKYNAPK R VVR +N V CQ++ + + GD ++ A S EL
Sbjct 25 EGKTDYYARKRLIAQAKNKYNAPKYRLVVRFSNRFVTCQIVSSRVNGDYVLAHAHSSELP 84
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEVLRGSAGP 138
RYGIK GL N++AAYATGL +ARR L + GLAD+++G+ +P G E+ + E + P
Sbjct 85 RYGIKWGLANWTAAYATGLLVARRALAKVGLADKYEGVTEPEG-EFELTEAIEDGPRP 141
> Hs14591909
Length=297
Score = 128 bits (321), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 63/111 (56%), Positives = 88/111 (79%), Gaps = 0/111 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY AR+RL++QDKNKYN PK R +VRVTN ++CQ+ YA ++GD +VC+A + EL
Sbjct 25 EGKTDYYARKRLVIQDKNKYNTPKYRMIVRVTNRDIICQIAYARIEGDMIVCAAYAHELP 84
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEV 131
+YG+KVGLTNY+AAY TGL LARRLL + G+ ++G + +G+EY++E +
Sbjct 85 KYGVKVGLTNYAAAYCTGLLLARRLLNRFGMDKIYEGQVEVTGDEYNVESI 135
> 7289495
Length=319
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 64/134 (47%), Positives = 85/134 (63%), Gaps = 20/134 (14%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY AR+RL QDKNKYN PK R +VR++N + Q+ YA ++GDR+VC+A S EL
Sbjct 25 EGKTDYYARKRLTFQDKNKYNTPKYRLIVRLSNKDITVQIAYARIEGDRVVCAAYSHELP 84
Query 81 RYGI--------------------KVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEK 120
+YGI KVGLTNY+AAY TGL +ARR+L + GL + G +
Sbjct 85 KYGIQVSFPTAFWRTQLDGPLYLFKVGLTNYAAAYCTGLLVARRVLNKLGLDSLYAGCTE 144
Query 121 PSGEEYHIEEVLRG 134
+GEE+++E V G
Sbjct 145 VTGEEFNVEPVDDG 158
> At5g39740
Length=301
Score = 125 bits (313), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 60/109 (55%), Positives = 81/109 (74%), Gaps = 0/109 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
+GKTDY AR RLI QDKNKYN PK RFVVR TN ++ Q++ A++ GD + SA + EL
Sbjct 25 DGKTDYRARIRLINQDKNKYNTPKYRFVVRFTNKDIVAQIVSASIAGDIVKASAYAHELP 84
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIE 129
+YG+ VGLTNY+AAY TGL LARR+LK + DE++G + +GE++ +E
Sbjct 85 QYGLTVGLTNYAAAYCTGLLLARRVLKMLEMDDEYEGNVEATGEDFSVE 133
> Hs17438041
Length=209
Score = 121 bits (303), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 58/121 (47%), Positives = 83/121 (68%), Gaps = 4/121 (3%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
+GKT+Y A + L++QDKNKY PK R +VRVTN ++CQ+ YA + D +VC+A + EL
Sbjct 56 KGKTEYHAWKHLVIQDKNKYKTPKYRMIVRVTNRDIICQIAYAHIDRDIIVCAAYAHELP 115
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEVLRGSAGPSN 140
+YG+K GLTNY AAY TGL LA RLL + G+ ++G + +G+EY++E + GP
Sbjct 116 KYGMKDGLTNYVAAYCTGLLLACRLLNRFGMDKIYEGQVEVTGDEYNVESI----DGPPG 171
Query 141 A 141
A
Sbjct 172 A 172
> Hs20482081
Length=195
Score = 120 bits (301), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 54/111 (48%), Positives = 80/111 (72%), Gaps = 0/111 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY AR+RL++QDKNKYN PK +V V N ++CQ+ Y ++G+ +V +A + EL
Sbjct 14 EGKTDYYARKRLVIQDKNKYNTPKYWMIVHVINRYIICQIAYGCIEGNVIVRAAHAHELP 73
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEV 131
+YG+KVGLTNY+AAY T L LA RLL + G+ ++G + + +EY++E +
Sbjct 74 KYGVKVGLTNYAAAYCTCLLLACRLLNRFGMDKIYEGQVEATSDEYNVESI 124
> YPL131w
Length=297
Score = 114 bits (286), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 53/96 (55%), Positives = 71/96 (73%), Gaps = 0/96 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY R+RL+ Q K KYN PK R VVR TN ++CQ++ +T+ GD ++ +A S EL
Sbjct 25 EGKTDYYQRKRLVTQHKAKYNTPKYRLVVRFTNKDIICQIISSTITGDVVLAAAYSHELP 84
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFK 116
RYGI GLTN++AAYATGL +ARR L++ GL + +K
Sbjct 85 RYGITHGLTNWAAAYATGLLIARRTLQKLGLDETYK 120
> Hs20473693
Length=451
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 53/111 (47%), Positives = 78/111 (70%), Gaps = 2/111 (1%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY A++RL++QDKNKY+ PK R +V VTN ++ ++GD +VC+A + EL
Sbjct 300 EGKTDYYAQKRLVMQDKNKYSTPKYRMIVCVTNRDII--YCLGHIEGDMIVCAAYAHELP 357
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEV 131
+YG+KVGLTNY+AAY GL L R+L + G+ ++G K +G+EY +E +
Sbjct 358 KYGVKVGLTNYAAAYCPGLLLTHRILSRFGMDKIYEGQVKVTGDEYDVESI 408
> Hs22053271_1
Length=329
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 50/108 (46%), Positives = 75/108 (69%), Gaps = 0/108 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY A + L++QDKNK N K R ++ V N+ +C++ YA ++ D +VC+A + EL
Sbjct 97 EGKTDYYAWKHLVVQDKNKSNTHKYRMIICVINTDTICEMAYAHIEWDMIVCAAYAHELP 156
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHI 128
+YG+KVGLTN +AA TGL LA RLL + G+ +KG + + +EY++
Sbjct 157 KYGVKVGLTNDAAACCTGLLLACRLLSRFGMDKIYKGQVEVTRDEYNV 204
> ECU06g0900i
Length=283
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/99 (51%), Positives = 65/99 (65%), Gaps = 0/99 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY R +I QD N + K R VVR+T SRV+CQ++ A + GDR++ ADS+EL
Sbjct 19 EGKTDYKHRTNMIRQDSNSCGSVKHRLVVRITGSRVICQIVAAYMDGDRVMVYADSRELE 78
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLE 119
RYGI G TNYSAAYATG + RR L L + ++ E
Sbjct 79 RYGINFGATNYSAAYATGFLIGRRALTAMSLDEVYRPKE 117
> Hs20534945
Length=123
Score = 100 bits (249), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 45/78 (57%), Positives = 59/78 (75%), Gaps = 0/78 (0%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKT Y AR+ L +QDKNKYN PK R VV +TN ++ Q+ YA ++GD +VC+A + EL
Sbjct 25 EGKTGYYARKYLEIQDKNKYNTPKYRIVVHLTNRDIISQIAYARIEGDMIVCAAYAHELP 84
Query 81 RYGIKVGLTNYSAAYATG 98
+YG+KVGLTNY+A Y TG
Sbjct 85 KYGVKVGLTNYAAEYCTG 102
> Hs22057807
Length=247
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 42/85 (49%), Positives = 63/85 (74%), Gaps = 0/85 (0%)
Query 47 FVVRVTNSRVLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLXLARRLL 106
+VRVTN ++CQ+ YA ++GD +VC+A + EL +YG+KVG TNY+AAY TGL LARRLL
Sbjct 1 MIVRVTNRDIICQIAYAHIEGDMIVCTAYAHELPKYGVKVGPTNYAAAYCTGLPLARRLL 60
Query 107 KQKGLADEFKGLEKPSGEEYHIEEV 131
+ G+ ++G + +G+EY +E +
Sbjct 61 SRFGMDKIYEGQVEVTGDEYIVESI 85
> Hs18576820
Length=133
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 60/87 (68%), Gaps = 0/87 (0%)
Query 43 PKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLXLA 102
PK R + +TN+ +C + YA ++GD ++C + EL ++G+KVGLTNY+AA+ GL LA
Sbjct 2 PKYRMIAHITNTESVCHIAYACMEGDMIMCVPYAHELPKHGVKVGLTNYAAAHCPGLLLA 61
Query 103 RRLLKQKGLADEFKGLEKPSGEEYHIE 129
RLL + G+ ++G + +G+EY++E
Sbjct 62 CRLLNRSGMGKIYEGQVEVTGDEYNVE 88
> Hs17441351
Length=131
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 62/110 (56%), Gaps = 17/110 (15%)
Query 5 TKRFLSASRSSTGGAFEGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYAT 64
TK + + ++GK DY A++RL D + VT+ ++C + YA
Sbjct 9 TKAYFKRHQVKFRRQWDGKIDYYAQKRL---DDS------------VTD--IICHIAYAC 51
Query 65 LQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLXLARRLLKQKGLADE 114
++GD +VC A + EL +YG+KVGLTNY+AAY TGL L RLL + DE
Sbjct 52 IEGDMIVCIAYAHELPKYGVKVGLTNYAAAYCTGLLLDHRLLNRFVEEDE 101
> Hs17463429
Length=196
Score = 71.2 bits (173), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 37/84 (44%), Positives = 60/84 (71%), Gaps = 0/84 (0%)
Query 48 VVRVTNSRVLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLXLARRLLK 107
+V VT R++CQ+ Y ++GD +VC+A + +L +YG+ VGLTNY+AAY TGL LARRLL
Sbjct 2 IVHVTKRRIICQIAYVRIEGDMIVCAAYAHKLPKYGVNVGLTNYAAAYCTGLLLARRLLN 61
Query 108 QKGLADEFKGLEKPSGEEYHIEEV 131
+ + ++ + +G+EY++E +
Sbjct 62 RFEVDKIYESQVEKTGDEYNVESI 85
> Hs17446861
Length=230
Score = 71.2 bits (173), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 43/108 (39%), Positives = 66/108 (61%), Gaps = 14/108 (12%)
Query 21 EGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELT 80
EGKTDY A++ L D + +N ++CQ+ YA +GD +VC+A EL
Sbjct 25 EGKTDYYAQKHL---DDSSWN-----------KQSIICQIAYARTEGDMIVCTAYVHELP 70
Query 81 RYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHI 128
+YG+KV LTNY+AAY TGL L RRLL + + ++G + +G++Y++
Sbjct 71 KYGVKVDLTNYAAAYCTGLLLDRRLLNRFDMDKIYEGQVEATGDDYNV 118
> Hs20542597
Length=336
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 48/66 (72%), Gaps = 0/66 (0%)
Query 66 QGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEE 125
+G+ +VC+A + EL +Y +KVGLTNY+AAY TGL LA LL + G+ + + G + +G+E
Sbjct 178 EGEIIVCAAYAHELPKYDVKVGLTNYAAAYCTGLLLACSLLNRFGMDNIYDGQMEVTGDE 237
Query 126 YHIEEV 131
Y++E +
Sbjct 238 YNVENI 243
> Hs20471297
Length=395
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 43/66 (65%), Gaps = 0/66 (0%)
Query 66 QGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEE 125
+GD +VC+A + EL +YG+K GLTN +A Y T L LA RLL + G+ + +G+E
Sbjct 253 EGDMIVCTAYAHELPKYGMKFGLTNDAAMYCTVLLLAHRLLNKFGMEKICEHQVAVTGDE 312
Query 126 YHIEEV 131
Y++E +
Sbjct 313 YNVEGI 318
> Hs22045091
Length=319
Score = 54.3 bits (129), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 78 ELTRYGIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHIEEV 131
EL +YG+KVGLT+Y+ AY TGL LA RLL + G+ ++G + + +E+++E +
Sbjct 223 ELPKYGVKVGLTHYATAYCTGLLLACRLLSKSGMVKIYEGQVEVTRDEHNVENI 276
> Hs22050098
Length=247
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 58/120 (48%), Gaps = 16/120 (13%)
Query 8 FLSA--SRSSTGGAFEGKTDYAARRRLILQDKNKYNAPKCRFVVRVTNSRVLCQVMYATL 65
F+SA + ++GG+F + L L D+ + A +V +TN V C ++YA +
Sbjct 118 FISAVDEKETSGGSF--------HQNLPLNDETELEASHLMRIVDITNRDVNCYLVYAGI 169
Query 66 QGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLXLA-RRLLKQKGLADEFKGLEKPSGE 124
+GD +VC+A + +L RYG+ N + Y + + + LA +F P+ +
Sbjct 170 EGDMIVCTAHAHKLPRYGV-----NLAKLYCSTMKWPTENVFSTLHLATQFYEFSPPNSD 224
> Hs22043196_2
Length=415
Score = 32.3 bits (72), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 18/24 (75%), Gaps = 1/24 (4%)
Query 46 RFVVRVTNSRVL-CQVMYATLQGD 68
R +V VT +RV+ CQ+ YA +QGD
Sbjct 240 RMIVHVTPNRVIICQIAYACMQGD 263
> Hs22044614
Length=455
Score = 30.0 bits (66), Expect = 1.3, Method: Composition-based stats.
Identities = 13/24 (54%), Positives = 18/24 (75%), Gaps = 1/24 (4%)
Query 46 RFVVRVTNSRVL-CQVMYATLQGD 68
R +V VT +RV+ CQ+ YA +QGD
Sbjct 280 RMIVHVTPNRVIICQIAYACMQGD 303
> Hs8924244
Length=354
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 54 SRVLCQVMYATLQGDRLVCSADSQELTR 81
S+V+C+V + TLQGD L +A+ E R
Sbjct 189 SQVICEVAHVTLQGDPLRGTANLSEAIR 216
> HsM4758978
Length=503
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 53 NSRVLCQVMYATLQGDRLVCSADSQELTR 81
+S+V+C+V + TLQGD L +A+ E R
Sbjct 222 HSQVICEVAHVTLQGDPLRGTANLSETIR 250
> Hs18426911
Length=504
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 53 NSRVLCQVMYATLQGDRLVCSADSQELTR 81
+S+V+C+V + TLQGD L +A+ E R
Sbjct 223 HSQVICEVAHVTLQGDPLRGTANLSETIR 251
> At5g39790
Length=271
Score = 28.9 bits (63), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 48/106 (45%), Gaps = 14/106 (13%)
Query 29 RRRLI--LQDKNKYNAPKCRFVVR----VTNSRVLCQVMYATLQGDRLVCSADSQELTRY 82
R +L+ L + N+ N RF+ R +TN + +M +Q LV A+ E+
Sbjct 96 RSKLVKKLSEANQQN----RFLKRQEHEITNIKTELALMELEVQA--LVKLAE--EIANL 147
Query 83 GIKVGLTNYSAAYATGLXLARRLLKQKGLADEFKGLEKPSGEEYHI 128
GI G S Y L+R QK + ++ KG+E +E H+
Sbjct 148 GIPQGSRKISGKYIQSHLLSRLDAVQKKMKEQIKGVEAAQSKEVHV 193
> Hs22043418
Length=213
Score = 27.3 bits (59), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Query 60 VMYATLQG-DRLVCSADSQELTRYGIKVGLTNYSAAYATG 98
V+Y++L G +V SADS E + G K G+ +Y+ ATG
Sbjct 113 VLYSSLTGGPSVVTSADS-EAAQCGCKKGIDSYNKVIATG 151
Lambda K H
0.317 0.132 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40