bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4956_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
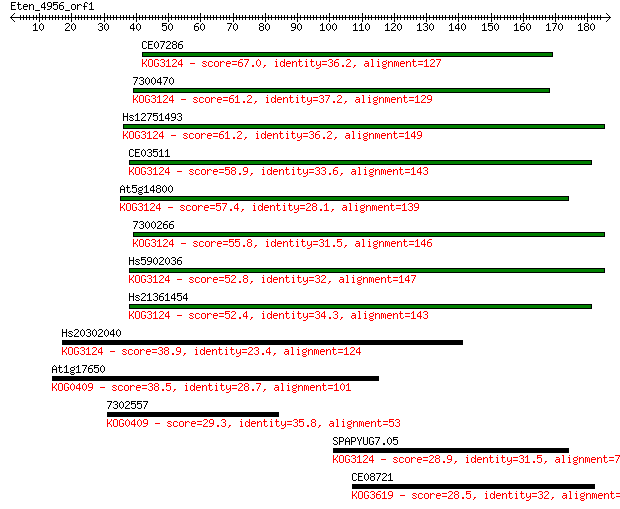
CE07286 67.0 2e-11
7300470 61.2 1e-09
Hs12751493 61.2 1e-09
CE03511 58.9 5e-09
At5g14800 57.4 1e-08
7300266 55.8 4e-08
Hs5902036 52.8 4e-07
Hs21361454 52.4 5e-07
Hs20302040 38.9 0.005
At1g17650 38.5 0.007
7302557 29.3 4.4
SPAPYUG7.05 28.9 5.7
CE08721 28.5 7.5
> CE07286
Length=299
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 46/133 (34%), Positives = 65/133 (48%), Gaps = 7/133 (5%)
Query 42 FLGGGKMARAMAASLVRAGLCLPADIWMSARTTTTYEYIRDLGY-GTGTNT-EVVERTQG 99
F+GGG MA A+ G ++I + +T + E R LGY TNT E++ER
Sbjct 19 FIGGGNMAAAIIKGCQNKGFTPKSNIVIGVQTEKSAEKWRQLGYKNVFTNTLEMLERYST 78
Query 100 GLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGVPL----ARLLKACDNVPVARIMPN 155
+ + VKP V EV+ R ++S GVPL A+L N + R+MPN
Sbjct 79 AIYVICVKPQVFEEVVSS-WPVNSRPEFIISVMAGVPLKVLNAKLPFVSGNTTIVRLMPN 137
Query 156 VLCSVGEGATCYC 168
V S+G GA+ C
Sbjct 138 VASSIGAGASTMC 150
> 7300470
Length=280
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 66/140 (47%), Gaps = 13/140 (9%)
Query 39 RAGFLGGGKMARAMAASLVRAGLCLPADIWMSARTTTTYEYIRDLGYGT-GTNTEVVERT 97
+ GF+GGG MA A+ + LVR G+ + + +S + RDLG T N V+E +
Sbjct 7 KIGFIGGGNMAYAIGSGLVRCGIVKASQVQVSGPHIENLQRWRDLGAVTCDDNCMVLEHS 66
Query 98 QGGLLFLTVKPAV-----ALEVLQEVRHAVDRVALVVSACVGVPLARLLKA-----CDNV 147
++F+ VKP + A + V A D LVVS G L L +A +
Sbjct 67 D--IVFICVKPHMLTPCAAQLKYKHVPSAKDASKLVVSVLAGTSLETLEEAFSFMGSSEL 124
Query 148 PVARIMPNVLCSVGEGATCY 167
V R MPN VGEG T Y
Sbjct 125 KVIRTMPNTSMQVGEGCTVY 144
> Hs12751493
Length=274
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 54/151 (35%), Positives = 72/151 (47%), Gaps = 4/151 (2%)
Query 36 STLRAGFLGGGKMARAMAASLVRAGLCLPADIWMSARTTTTYEYIRDLG-YGTGTNTEVV 94
S R GF+G G+MA A+A L+RAG I SA T + + LG T +N EV+
Sbjct 7 SPRRVGFVGAGRMAGAIAQGLIRAGKVEAQHILASAPTDRNLCHFQALGCRTTHSNQEVL 66
Query 95 ERTQGGLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGVPLARLLKAC-DNVPVARIM 153
+ L+ KP V VL EV V ++VS GV L+ L + N V R++
Sbjct 67 QSCL--LVIFATKPHVLPAVLAEVAPVVTTEHILVSVAAGVSLSTLEELLPPNTRVLRVL 124
Query 154 PNVLCSVGEGATCYCLSPCAESAHALLLQAL 184
PN+ C V EGA S+ LLQ L
Sbjct 125 PNLPCVVQEGAIVMARGRHVGSSETKLLQHL 155
> CE03511
Length=279
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 48/152 (31%), Positives = 77/152 (50%), Gaps = 11/152 (7%)
Query 38 LRAGFLGGGKMARAMAASLVRAGLCLPADIWMSA--RTTTTYEYIRDLGYG-TGTNTEVV 94
++ GF+G GKMA+A+A L+ +G +I S+ R + + LG T N EVV
Sbjct 1 MKIGFIGAGKMAQALARGLINSGRITADNIIASSPKRDEVFLDQCKALGLNTTHDNAEVV 60
Query 95 ERTQGGLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGVPLARLLKACDNVP-VARIM 153
+++ ++FL VKP +V E+ A+ + LVVS +G+ + + V R+M
Sbjct 61 QKSD--VVFLAVKPVHVSKVASEIAPALSKEHLVVSIALGITIRNIESLLPTKSRVVRVM 118
Query 154 PNVLCSVGEGATCYCL-SPC----AESAHALL 180
PN V GA+ + + S C AE+ LL
Sbjct 119 PNTPSVVRAGASAFAMGSACRDGDAETVEKLL 150
> At5g14800
Length=276
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 69/144 (47%), Gaps = 10/144 (6%)
Query 35 SSTLRAGFLGGGKMARAMAASLVRAGLCLPADIWMSARTTTTYEYIRDLGYGTGTNT--- 91
+ + + GF+G GKMA ++A +V +G+ P I + + RD+ G N
Sbjct 8 AESFKVGFIGAGKMAESIARGVVASGVLPPNRICTAVHSNLNR---RDVFESFGVNVFST 64
Query 92 --EVVERTQGGLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGVPLARLLKACDNVPV 149
EVV+ + ++ +VKP V + + E++ + + ++VS G+ L L +
Sbjct 65 SEEVVKESD--VVIFSVKPQVVKKAVTELKSKLSKNKILVSVAAGIKLNDLQEWSGQDRF 122
Query 150 ARIMPNVLCSVGEGATCYCLSPCA 173
R+MPN +VGE A+ L A
Sbjct 123 IRVMPNTPAAVGEAASVMSLGTGA 146
> 7300266
Length=273
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 46/150 (30%), Positives = 73/150 (48%), Gaps = 7/150 (4%)
Query 39 RAGFLGGGKMARAMAASLVRAGLCLPADIWMSARTTT--TYEYIRDLGYGTG-TNTEVVE 95
+ GFLGGG MA+A+A + AGL P + S + + + LG T N VV+
Sbjct 6 KIGFLGGGNMAKALAKGFLAAGLAKPNTLIASVHPADKLSLQSFQSLGVETVIKNAPVVQ 65
Query 96 RTQGGLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGVPLARLLKA-CDNVPVARIMP 154
Q ++F++VKP V VL E++ + L +S +G+ L+ + + V R+MP
Sbjct 66 --QSDVVFVSVKPQVVPSVLSEIQ-PLSSGKLFLSVAMGITLSTIESSLSPQARVIRVMP 122
Query 155 NVLCSVGEGATCYCLSPCAESAHALLLQAL 184
N+ V G + + A A A + Q L
Sbjct 123 NLPAVVCSGCSVFVRGSKATDADADITQKL 152
> Hs5902036
Length=319
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 47/154 (30%), Positives = 71/154 (46%), Gaps = 9/154 (5%)
Query 38 LRAGFLGGGKMARAMAASLVRAGLCLPADIWMSARTTT--TYEYIRDLGYG-TGTNTEVV 94
+ GF+G G++A A+A AG+ I S+ T +R +G T N E V
Sbjct 1 MSVGFIGAGQLAFALAKGFTAAGVLAAHKIMASSPDMDLATVSALRKMGVKLTPHNKETV 60
Query 95 ERTQGGLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGVPLARL---LKACDNVP-VA 150
+ + +LFL VKP + +L E+ ++ +VVS GV ++ + L A P V
Sbjct 61 QHSD--VLFLAVKPHIIPFILDEIGADIEDRHIVVSCAAGVTISSIEKKLSAFRPAPRVI 118
Query 151 RIMPNVLCSVGEGATCYCLSPCAESAHALLLQAL 184
R M N V EGAT Y A+ L++ L
Sbjct 119 RCMTNTPVVVREGATVYATGTHAQVEDGRLMEQL 152
> Hs21361454
Length=320
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 49/150 (32%), Positives = 70/150 (46%), Gaps = 14/150 (9%)
Query 38 LRAGFLGGGKMARAMAASLVRAGLCLPADIWMSARTTT--TYEYIRDLGYG-TGTNTEVV 94
+ GF+G G++A A+A AG+ I S+ T +R +G T +N E V
Sbjct 1 MSVGFIGAGQLAYALARGFTAAGILSAHKIIASSPEMNLPTVSALRKMGVNLTRSNKETV 60
Query 95 ERTQGGLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGVPLA---RLLKACDNVP-VA 150
+ + +LFL VKP + +L E+ V +VVS GV ++ + L A P V
Sbjct 61 KHSD--VLFLAVKPHIIPFILDEIGADVQARHIVVSCAAGVTISSVEKKLMAFQPAPKVI 118
Query 151 RIMPNVLCSVGEGATCYCLSPCAESAHALL 180
R M N V EGAT Y A HAL+
Sbjct 119 RCMTNTPVVVQEGATVY-----ATGTHALV 143
> Hs20302040
Length=250
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 58/125 (46%), Gaps = 3/125 (2%)
Query 17 TQKLSLRAILIFSRMSERSSTLRAGFLGGGKMARAMAASLVRAGLCLPADIWMSARTTTT 76
+ S R + I S S + G +GGG + + +A +L++ G + +S R T
Sbjct 56 NKNQSSRHLSIGSLNSATPEEFKVGIIGGGHLGKQLAGTLLQLGPIPAESLRISTRRPET 115
Query 77 YEYIRDLGYG-TGTNTEVVERTQGGLLFLTVKPAVALEVLQEVRHAVDRVALVVSACVGV 135
++ LG N ++V + ++FL P+ + E+ ++++ ++V S +
Sbjct 116 LGELQKLGIKCFYHNADLV--SWADVIFLCCLPSQLPNICVEIYTSLEKASIVYSFVAAI 173
Query 136 PLARL 140
PL RL
Sbjct 174 PLPRL 178
> At1g17650
Length=670
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 29/101 (28%), Positives = 49/101 (48%), Gaps = 6/101 (5%)
Query 14 SFRTQKLSLRAILIFSRMSERSSTLRAGFLGGGKMARAMAASLVRAGLCLPADIWMSART 73
S ++ S R I++ RM++ T+ GFLG G M MA +L++AG D+ + RT
Sbjct 331 SLQSTTPSTRGIVV--RMADELGTVSIGFLGMGIMGSPMAQNLIKAG----CDVTVWNRT 384
Query 74 TTTYEYIRDLGYGTGTNTEVVERTQGGLLFLTVKPAVALEV 114
+ + + LG ++ E V T + P A++V
Sbjct 385 KSKCDPLVGLGAKYKSSPEEVTATCDLTFAMLADPESAIDV 425
> 7302557
Length=315
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 29/60 (48%), Gaps = 7/60 (11%)
Query 31 MSERSSTLRAGFLGGGKMARAMAASLVRAGLCL-------PADIWMSARTTTTYEYIRDL 83
MS + GF+G G M MA++L++AG L PA ++A+ T Y +L
Sbjct 13 MSTQGGAKNIGFVGLGNMGANMASNLIKAGHKLHVFDISKPACDGLAAKGATVYAKTSEL 72
> SPAPYUG7.05
Length=282
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 38/76 (50%), Gaps = 4/76 (5%)
Query 101 LLFLTVKPAVALEVLQ--EVRHAVDRVALVVSACVGVPLARLLKACD-NVPVARIMPNVL 157
+L L+ KP A +VL +++ A+ + L++S G ++ L D + V RIMPN
Sbjct 80 VLLLSCKPQAAEDVLNSPKMKEAL-KGKLILSILAGKTISSLQSMLDESTRVIRIMPNTA 138
Query 158 CSVGEGATCYCLSPCA 173
+ E + C P A
Sbjct 139 SRIRESMSVICPGPNA 154
> CE08721
Length=805
Score = 28.5 bits (62), Expect = 7.5, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 6/80 (7%)
Query 107 KPAVALEVLQEVRHAVDRVA---LVVSACVGVPLARLLKACDNVPVARIMPNVLCS--VG 161
KP + E +V A R+ V++ C + AR+LK D+ + R+ + C+
Sbjct 84 KPHLYNETYSQVSVAYGRLVGFESVLAQCSSIDAARVLKELDS-RIERLAASNGCTRVAS 142
Query 162 EGATCYCLSPCAESAHALLL 181
EG T C P +S HA L
Sbjct 143 EGITAVCSIPGIDSQHATKL 162
Lambda K H
0.324 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3022542264
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40