bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5089_orf2
Length=80
Score E
Sequences producing significant alignments: (Bits) Value
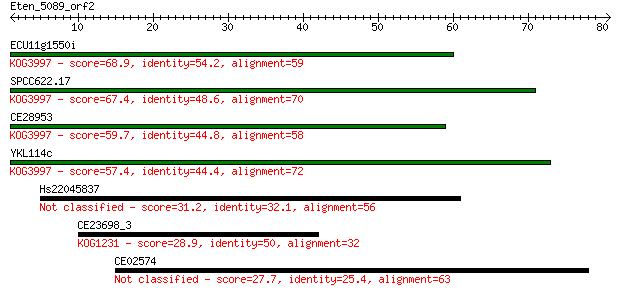
ECU11g1550i 68.9 2e-12
SPCC622.17 67.4 6e-12
CE28953 59.7 1e-09
YKL114c 57.4 6e-09
Hs22045837 31.2 0.51
CE23698_3 28.9 2.5
CE02574 27.7 5.1
> ECU11g1550i
Length=275
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/59 (54%), Positives = 42/59 (71%), Gaps = 1/59 (1%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDP 59
G +FL+ MH+NDSK L S DRHE +GKG +G F++IM + F ++PLILETPDP
Sbjct 202 GLEFLKAMHINDSKEALGSKKDRHESIGKGMIGEKAFRFIMNSKV-FDNIPLILETPDP 259
> SPCC622.17
Length=366
Score = 67.4 bits (163), Expect = 6e-12, Method: Composition-based stats.
Identities = 34/70 (48%), Positives = 47/70 (67%), Gaps = 2/70 (2%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPT 60
G K++ G HLNDSK+ L S D HE +G G LGL PF+ IM + +R+ +PL+LETP +
Sbjct 223 GAKYVSGWHLNDSKAPLGSNRDLHENIGLGFLGLEPFRLIM-NDSRWDGIPLVLETPAKS 281
Query 61 EGTIWKKEIK 70
WKKE++
Sbjct 282 PEQ-WKKEVE 290
> CE28953
Length=396
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 26/58 (44%), Positives = 42/58 (72%), Gaps = 1/58 (1%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPD 58
G+ +L+ +H+NDSK ++ S LDRHE +G+G +G A F+ +M + R +P+ILETP+
Sbjct 324 GWNYLKAIHINDSKGDVGSKLDRHEHIGQGKIGKAAFELLM-NDNRLDGIPMILETPE 380
> YKL114c
Length=367
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 47/73 (64%), Gaps = 4/73 (5%)
Query 1 GYKFLRGMHLNDSKSELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETP-DP 59
G+K+L +HLNDSK+ L + D HE LG+G+LG+ F+ I H + +P++LETP +
Sbjct 221 GFKYLSAVHLNDSKAPLGANRDLHERLGQGYLGIDVFRMIA-HSEYLQGIPIVLETPYEN 279
Query 60 TEGTIWKKEIKQM 72
EG + EIK M
Sbjct 280 DEG--YGNEIKLM 290
> Hs22045837
Length=252
Score = 31.2 bits (69), Expect = 0.51, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 4/60 (6%)
Query 5 LRGMHLNDSKSELSSGLDRHELLGK----GHLGLAPFKYIMQHQTRFKDMPLILETPDPT 60
+RG H +DS+ L S D ++ G++ P+ M+ QTR + P + T DP+
Sbjct 42 IRGHHYSDSRRSLESFRDHAAIIYPQHHCGNISTGPYTVRMEPQTRQQQQPGGVGTSDPS 101
> CE23698_3
Length=454
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 10 LNDSKSELSSGLDRHELLGK--GHLGLAPFKYIM 41
L D SE S LD HE+LG GH+G F I+
Sbjct 342 LADVISETRSDLDEHEILGTVVGHVGDGNFHVIL 375
> CE02574
Length=320
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/63 (25%), Positives = 29/63 (46%), Gaps = 0/63 (0%)
Query 15 SELSSGLDRHELLGKGHLGLAPFKYIMQHQTRFKDMPLILETPDPTEGTIWKKEIKQMYL 74
S LS L+R + L H+ L P + + +F+ + DP++ + IK+M+
Sbjct 13 SALSIPLNRSDFLENDHVELMPLEKDGELNDKFRQEMIFSHNLDPSDSKQLSESIKEMFK 72
Query 75 GRD 77
D
Sbjct 73 KTD 75
Lambda K H
0.319 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1165602088
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40