bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5112_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
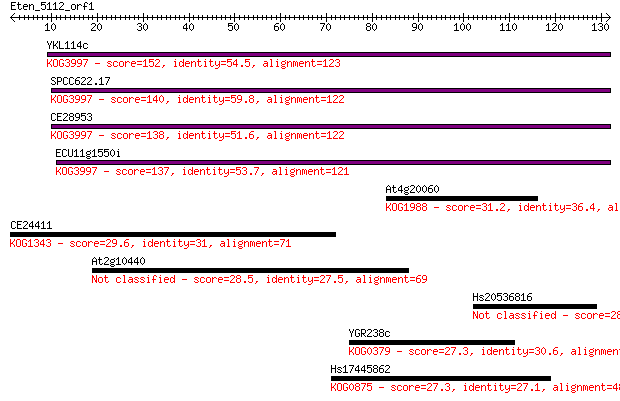
YKL114c 152 2e-37
SPCC622.17 140 8e-34
CE28953 138 3e-33
ECU11g1550i 137 5e-33
At4g20060 31.2 0.60
CE24411 29.6 1.6
At2g10440 28.5 3.7
Hs20536816 28.1 4.9
YGR238c 27.3 8.5
Hs17445862 27.3 8.5
> YKL114c
Length=367
Score = 152 bits (383), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 67/123 (54%), Positives = 93/123 (75%), Gaps = 1/123 (0%)
Query 9 VQVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGI 68
VLPHG Y IN+ANPD+ K E +Y + +DDL RCE LGI LYN+HPGST+ + + +
Sbjct 78 TDVLPHGQYFINLANPDREKAEKSYESFMDDLNRCEQLGIGLYNLHPGSTL-KGDHQLQL 136
Query 69 RNIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHL 128
+ +A+ +N+A K+TKFV +VLENMAG N+VGS DL+++I ++E+K R+GVC+DTCH
Sbjct 137 KQLASYLNKAIKETKFVKIVLENMAGTGNLVGSSLVDLKEVIGMIEDKSRIGVCIDTCHT 196
Query 129 FAA 131
FAA
Sbjct 197 FAA 199
> SPCC622.17
Length=366
Score = 140 bits (352), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 73/146 (50%), Positives = 92/146 (63%), Gaps = 25/146 (17%)
Query 10 QVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYN----IH----------- 54
QVL HGSYLIN+AN D+ K+E A+N +DDL+RCE LG+ LYN IH
Sbjct 57 QVLVHGSYLINMANADEQKREQAFNCFVDDLKRCERLGVGLYNFQYVIHSVLYLLIFYPK 116
Query 55 ---------PGSTVGECSKEEGIRNIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFED 105
PGST C+KEEGI N+A +NRAH++TK V++V ENMAGQ N +G F+D
Sbjct 117 RTNSFDLCSPGSTAS-CTKEEGINNLAECINRAHEETKSVIIVTENMAGQGNCLGGTFDD 175
Query 106 LRDIIELVENKDRVGVCLDTCHLFAA 131
+ ++N DR VCLDTCH FAA
Sbjct 176 FAALKSKIKNLDRWRVCLDTCHTFAA 201
> CE28953
Length=396
Score = 138 bits (347), Expect = 3e-33, Method: Composition-based stats.
Identities = 63/122 (51%), Positives = 88/122 (72%), Gaps = 0/122 (0%)
Query 10 QVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIR 69
Q++PHGSYL+N +P+ K E + A+LD+ QR E LGI +YN HPGSTVG+C KEE +
Sbjct 181 QIVPHGSYLMNAGSPEAEKLEKSRLAMLDECQRAEKLGITMYNFHPGSTVGKCEKEECMT 240
Query 70 NIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHLF 129
IA ++ + T+ +++VLE MAGQ N +G FE+L+ II+ V+ K RVGVC+DTCH+F
Sbjct 241 TIAETIDFVVEKTENIILVLETMAGQGNSIGGTFEELKFIIDKVKVKSRVGVCIDTCHIF 300
Query 130 AA 131
A
Sbjct 301 AG 302
> ECU11g1550i
Length=275
Score = 137 bits (345), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 65/121 (53%), Positives = 90/121 (74%), Gaps = 3/121 (2%)
Query 11 VLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIRN 70
++PHGSYLIN NPD + + L+DDL+RC++LGI +YN+HPGS+ G+ + E +R+
Sbjct 63 IVPHGSYLINFGNPDLV--DKSMECLVDDLERCKSLGIGMYNMHPGSSKGK-GRAECLRS 119
Query 71 IAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHLFA 130
I VN A + V V++ENMAGQ +VVGS FE+LRDII +E+++R+GVCLDTCHLF
Sbjct 120 IGHHVNTALSRVQGVTVLIENMAGQGSVVGSSFEELRDIIGGIEDRERIGVCLDTCHLFG 179
Query 131 A 131
A
Sbjct 180 A 180
> At4g20060
Length=1134
Score = 31.2 bits (69), Expect = 0.60, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 83 KFVVVVLENMAGQKNVVGSKFEDLRDIIELVEN 115
KF++V LEN+ G N++ +E ++ I E V +
Sbjct 510 KFLIVFLENLEGDDNLLSEIYEKVKHITEFVSS 542
> CE24411
Length=517
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 31/74 (41%), Gaps = 14/74 (18%)
Query 1 AFERPPGQVQVLPHGSYLI---NVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGS 57
AF RPPG + G NVA KA +N L+ D Y++H G+
Sbjct 130 AFIRPPGHHAMPDEGCGFCIFNNVAIAAKAAIQNGQKVLIVD-----------YDVHAGN 178
Query 58 TVGECSKEEGIRNI 71
EC ++ G N+
Sbjct 179 GTQECVEQMGEGNV 192
> At2g10440
Length=935
Score = 28.5 bits (62), Expect = 3.7, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 36/72 (50%), Gaps = 3/72 (4%)
Query 19 INVANPDKAKQENAY---NALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIRNIAAAV 75
IN+ PD + Q ++Y ++L ++ + G+K+ NI PG + + KE R + V
Sbjct 733 INIVPPDMSSQIDSYEQLSSLESEVVSTTSSGLKVNNIAPGYALLQEIKETNGRLVETVV 792
Query 76 NRAHKDTKFVVV 87
+D+ +V
Sbjct 793 EICDEDSLGTIV 804
> Hs20536816
Length=557
Score = 28.1 bits (61), Expect = 4.9, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 8/35 (22%)
Query 102 KFEDLRDIIE--------LVENKDRVGVCLDTCHL 128
KF+ + DIIE L++ KD+ GV CHL
Sbjct 510 KFDSVEDIIEHYKNFPIILIDGKDKTGVHRKQCHL 544
> YGR238c
Length=882
Score = 27.3 bits (59), Expect = 8.5, Method: Composition-based stats.
Identities = 11/36 (30%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 75 VNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDII 110
+ A KD+ F L+N+ QK + S+ DL++++
Sbjct 591 IKEASKDSNFQTARLKNLEIQKTFLESRINDLKNLL 626
> Hs17445862
Length=146
Score = 27.3 bits (59), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 71 IAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKF-EDLRDIIELVENKDR 118
+ ++ H +F+ + + + +K+++G KF +DLR +IE EN +
Sbjct 53 VDGGLSNPHSSKRFLGLSIPHKVHRKHIMGQKFADDLRYLIEEDENASK 101
Lambda K H
0.318 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40