bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5202_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
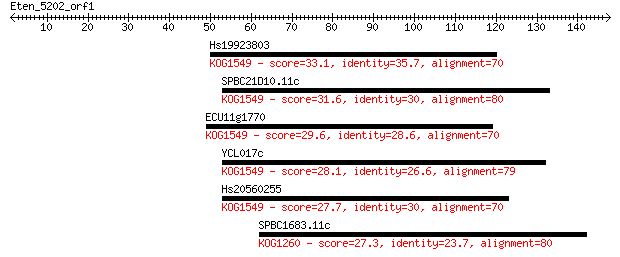
Hs19923803 33.1 0.20
SPBC21D10.11c 31.6 0.56
ECU11g1770 29.6 2.0
YCL017c 28.1 6.8
Hs20560255 27.7 8.2
SPBC1683.11c 27.3 9.9
> Hs19923803
Length=445
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 35/72 (48%), Gaps = 3/72 (4%)
Query 50 QWLPGTIHISIWGASGFEIANALSEEERLFVSAGVSCH--ECKTASGTLLAMGHLDEWAV 107
Q LP T + SI G + L++ L S G +CH S LL+ G + A
Sbjct 353 QRLPNTCNFSIRGPR-LQGHVVLAQCRVLMASVGAACHSDHGDQPSPVLLSYGVPFDVAR 411
Query 108 GSLRISVGRWTS 119
+LR+SVGR T+
Sbjct 412 NALRLSVGRSTT 423
> SPBC21D10.11c
Length=498
Score = 31.6 bits (70), Expect = 0.56, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 39/81 (48%), Gaps = 4/81 (4%)
Query 53 PGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAVGSLR 111
PG ++IS G + L + + +S+G +C S L A+G DE A S+R
Sbjct 392 PGCVNISFNYVEGESLLMGL---KNIALSSGSACTSASLEPSYVLRAIGQSDENAHSSIR 448
Query 112 ISVGRWTSPFEAADAARRVAK 132
+GR+T+ E A V++
Sbjct 449 FGIGRFTTEAEIDYAIENVSR 469
> ECU11g1770
Length=432
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 4/71 (5%)
Query 49 QQWLPGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAV 107
++ PG +++S G + L + + +S+G +C S L A+G DE A
Sbjct 323 EKGFPGCVNVSFPFVEGESLLMHLKD---IALSSGSACTSASLEPSYVLRALGRDDELAH 379
Query 108 GSLRISVGRWT 118
S+R +GR+T
Sbjct 380 SSIRFGIGRFT 390
> YCL017c
Length=497
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 38/80 (47%), Gaps = 4/80 (5%)
Query 53 PGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAVGSLR 111
PG +++S G + AL + + +S+G +C S L A+G D A S+R
Sbjct 391 PGCVNVSFAYVEGESLLMALRD---IALSSGSACTSASLEPSYVLHALGKDDALAHSSIR 447
Query 112 ISVGRWTSPFEAADAARRVA 131
+GR+++ E + V+
Sbjct 448 FGIGRFSTEEEVDYVVKAVS 467
> Hs20560255
Length=457
Score = 27.7 bits (60), Expect = 8.2, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 36/71 (50%), Gaps = 4/71 (5%)
Query 53 PGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAVGSLR 111
PG I++S G + AL + + +S+G +C S L A+G ++ A S+R
Sbjct 351 PGCINLSFAYVEGESLLMALKD---VALSSGSACTSASLEPSYVLRAIGTDEDLAHSSIR 407
Query 112 ISVGRWTSPFE 122
+GR+T+ E
Sbjct 408 FGIGRFTTEEE 418
> SPBC1683.11c
Length=518
Score = 27.3 bits (59), Expect = 9.9, Method: Composition-based stats.
Identities = 19/80 (23%), Positives = 36/80 (45%), Gaps = 13/80 (16%)
Query 62 GASGFEIANALSEEERLFVSAGVSCHECKTASGTLLAMGHLDEWAVGSLRISVGRWTSPF 121
G GF + + + FV +GV AM HLD+ A+G R +VG+ +
Sbjct 169 GDMGFGSVTSTMKMTKRFVESGV-------------AMIHLDDLAIGLKRFTVGQGRTVV 215
Query 122 EAADAARRVAKLLVSHQLLQ 141
++ RR+ + + +++
Sbjct 216 PTSEYLRRLTAVRLQFDIMK 235
Lambda K H
0.319 0.130 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40