bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5214_orf2
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
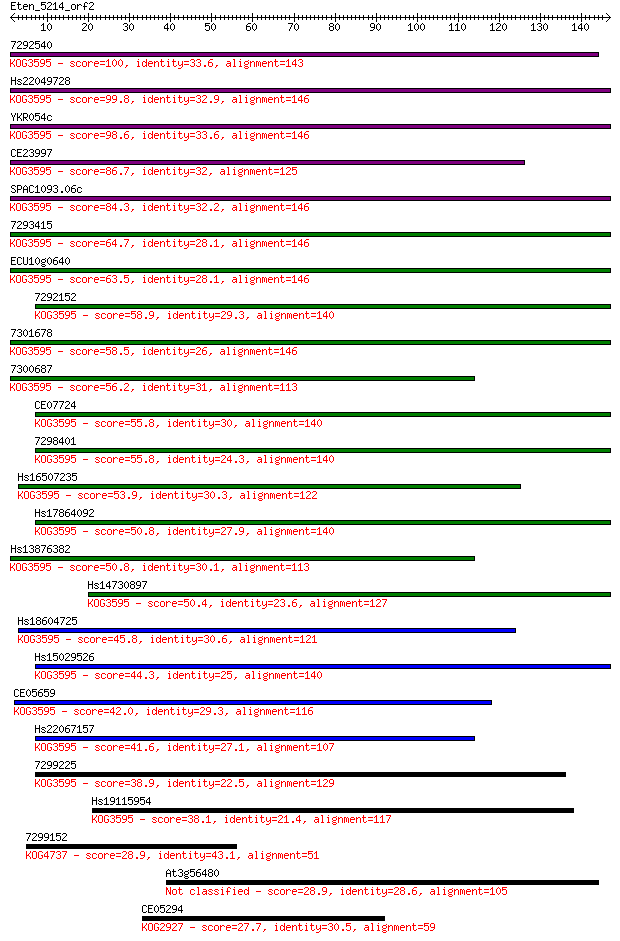
7292540 100 1e-21
Hs22049728 99.8 2e-21
YKR054c 98.6 4e-21
CE23997 86.7 2e-17
SPAC1093.06c 84.3 8e-17
7293415 64.7 6e-11
ECU10g0640 63.5 1e-10
7292152 58.9 3e-09
7301678 58.5 4e-09
7300687 56.2 2e-08
CE07724 55.8 2e-08
7298401 55.8 3e-08
Hs16507235 53.9 1e-07
Hs17864092 50.8 8e-07
Hs13876382 50.8 9e-07
Hs14730897 50.4 1e-06
Hs18604725 45.8 3e-05
Hs15029526 44.3 8e-05
CE05659 42.0 4e-04
Hs22067157 41.6 6e-04
7299225 38.9 0.003
Hs19115954 38.1 0.007
7299152 28.9 3.7
At3g56480 28.9 3.7
CE05294 27.7 8.1
> 7292540
Length=4680
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 48/145 (33%), Positives = 96/145 (66%), Gaps = 5/145 (3%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKIL-NFDC 59
K+ L E+++MANPP++ K+A+E++ +L+ GE W+ R ++ +FI+ I+ NF
Sbjct 3270 KQQLVEVRTMANPPSVVKLALESICLLL---GENATDWKSIRAVIMRENFINSIVSNFGT 3326
Query 60 TSVSSATRRRIQTKFLTG-DWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEI 118
+++ R ++++K+L+ D++ E++N+AS A GP+ +W + +++ + ++VEPL+ E+
Sbjct 3327 ENITDDVREKMKSKYLSNPDYNFEKVNRASMACGPMVKWAIAQIEYADMLKRVEPLREEL 3386
Query 119 AQLETEAQENEANLQEQQSLITQLE 143
LE +A N A+ +E + L+ QLE
Sbjct 3387 RSLEEQADVNLASAKETKDLVEQLE 3411
> Hs22049728
Length=2274
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 48/147 (32%), Positives = 90/147 (61%), Gaps = 4/147 (2%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCT 60
K+ L E++SMANPPA K+A+E++ +L+ GE W+ R I+ +FI I+NF
Sbjct 929 KQHLVEVRSMANPPAAVKLALESICLLL---GESTTDWKQIRSIIMRENFIPTIVNFSAE 985
Query 61 SVSSATRRRIQTKFLTG-DWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEIA 119
+S A R +++ +++ ++ E +N+AS A GP+ +W + + + + ++VEPL+ E+
Sbjct 986 EISDAIREKMKKNYMSNPSYNYEIVNRASLACGPMVKWAIAQLNYADMLKRVEPLRNELQ 1045
Query 120 QLETEAQENEANLQEQQSLITQLEGKL 146
+LE +A++N+ E + +I LE +
Sbjct 1046 KLEDDAKDNQQKANEVEQMIRDLEASI 1072
> YKR054c
Length=4092
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 49/148 (33%), Positives = 90/148 (60%), Gaps = 5/148 (3%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCT 60
K+ L E++SM NPP+ K+ +EAV ++ G + +W D ++ ++ DFI I+++D T
Sbjct 3117 KQQLTEIRSMVNPPSGVKIVMEAVCAIL---GYQFSNWRDIQQFIRKDDFIHNIVHYDTT 3173
Query 61 -SVSSATRRRIQTKFLTG-DWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEI 118
+ R+ ++ +FL+ ++ E IN+AS+A GPL QWV + + F + E V+PL+ E+
Sbjct 3174 LHMKPQIRKYMEEEFLSDPNFTYETINRASKACGPLYQWVNAQINFSKVLENVDPLRQEM 3233
Query 119 AQLETEAQENEANLQEQQSLITQLEGKL 146
++E E+ + +ANL + + LE +
Sbjct 3234 KRIEFESLKTKANLLAAEEMTQDLEASI 3261
> CE23997
Length=4568
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 40/126 (31%), Positives = 76/126 (60%), Gaps = 4/126 (3%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEK-PVSWEDARKILKATDFISKILNFDC 59
K L E+KSM++PP K+ +EA+ +L+ GE W+ R+++ DF+++IL FD
Sbjct 3244 KSQLVEVKSMSSPPVTVKLTLEAICILL---GENVGTDWKAIRQVMMKDDFMTRILQFDT 3300
Query 60 TSVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEIA 119
++ ++++ DW+ +++N+AS A GP+ +W + + + + +VEPL+ E+
Sbjct 3301 ELLTPEILKQMEKYIQNPDWEFDKVNRASVACGPMVKWARAQLLYSTMLHKVEPLRNELK 3360
Query 120 QLETEA 125
+LE EA
Sbjct 3361 RLEQEA 3366
> SPAC1093.06c
Length=1889
Score = 84.3 bits (207), Expect = 8e-17, Method: Composition-based stats.
Identities = 47/148 (31%), Positives = 85/148 (57%), Gaps = 5/148 (3%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDC- 59
K L EL+S++ PP ++ +E V L+ G W++ +++LK DFI KILN++
Sbjct 908 KAHLIELRSLSRPPMAIRITMEVVCKLL---GFSATDWKNVQQLLKRDDFIPKILNYNLE 964
Query 60 TSVSSATRRRIQTKFLTGD-WDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEI 118
+S RR+I+ + + + + +N+AS+A GPL W++S + + E++EPL +E+
Sbjct 965 KELSINLRRKIEQDYFSNPIFTFDSVNRASKACGPLLLWIKSICNYSKVLEKLEPLNSEV 1024
Query 119 AQLETEAQENEANLQEQQSLITQLEGKL 146
+L+ E + E +QE + L+ KL
Sbjct 1025 DRLKLEQKNAEECIQETIAACKDLDEKL 1052
> 7293415
Length=4081
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 41/146 (28%), Positives = 74/146 (50%), Gaps = 4/146 (2%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCT 60
K ++ELKS PPA+ + +EAV +L+ G KP +W A+ I+ +FI ++ +D
Sbjct 2727 KADINELKSFTTPPALVQFCMEAVCILL---GVKP-TWASAKAIMADINFIKRLFEYDKE 2782
Query 61 SVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEIAQ 120
+ T ++++ D+ + K S+ A ++ WV S KF + + VEP
Sbjct 2783 HMKEDTLKKVKKYIDHKDFVPAKFEKVSKVAKSMSMWVISMDKFSKVYKVVEPKIKRKEA 2842
Query 121 LETEAQENEANLQEQQSLITQLEGKL 146
E E +E L+++Q + +E K+
Sbjct 2843 AEAELKEVMTVLRQKQKELAAVEAKI 2868
> ECU10g0640
Length=3151
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 41/151 (27%), Positives = 77/151 (50%), Gaps = 11/151 (7%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLIT-----DAGEKPVSWEDARKILKATDFISKIL 55
K +L E+K M NPP + + VEAV L+ D G + W+ + +K DF+SK+L
Sbjct 2538 KSNLSEIKVMINPPEIVRSTVEAVFWLVEGRSKGDMGS--IEWKQLIQFMKREDFVSKVL 2595
Query 56 NFDCTSVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQ 115
+ + A + + E+ AS+A G L WV + K+ I ++ P++
Sbjct 2596 GCETLEIPDALEKLVSGPLFVH----EKALNASKACGSLFLWVMARYKYAKIMLEISPME 2651
Query 116 TEIAQLETEAQENEANLQEQQSLITQLEGKL 146
EIA LE + +++ ++E++ + ++E ++
Sbjct 2652 QEIAALEHDTKKSRKEIEEEERKLMEIEDRI 2682
> 7292152
Length=3868
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 41/151 (27%), Positives = 79/151 (52%), Gaps = 11/151 (7%)
Query 7 LKSMANPPAMAKVAVEAVAVLITDAGEK---PVS-------WEDARKILKATDFISKILN 56
+KSM NPP + K+ + AV V+ E+ P S W ++++L +F+ +
Sbjct 2633 VKSMKNPPPVIKLVMAAVCVIKGIPPERIPDPASGKMVQDYWGPSKRLLGEMNFLPGLKE 2692
Query 57 FDCTSVSSATRRRIQTKFLTG-DWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQ 115
FD ++ + +RI +F+ D+D + + KAS AA L QW+ + + + +++ V P +
Sbjct 2693 FDKDNIPTEIVKRIHKEFIPNKDFDPKVVAKASSAAKGLCQWIIAMMMYDEVAKVVAPKK 2752
Query 116 TEIAQLETEAQENEANLQEQQSLITQLEGKL 146
++A E E + L ++++L LE K+
Sbjct 2753 AKLAGAEKEYADTMEFLAQKRALALALEEKV 2783
> 7301678
Length=4820
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 38/146 (26%), Positives = 73/146 (50%), Gaps = 5/146 (3%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCT 60
K + E++S A PPA +V E VA+L G K ++W+ A+ ++ +F+ ++ DC
Sbjct 3711 KAQITEIRSFATPPAAVQVVCECVAIL---KGYKEINWKSAKGMMSDVNFLKSLMEMDCE 3767
Query 61 SVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEIAQ 120
+++ + + TG ++E + K S A L ++V + + F + ++V+P + +
Sbjct 3768 ALTQKQITQCRQHMKTG--NLEDMAKISVAGAGLLRFVRAVLGFFDVYKEVKPKKERLDF 3825
Query 121 LETEAQENEANLQEQQSLITQLEGKL 146
L E + L I +LE KL
Sbjct 3826 LVEEQEVQIKLLNHLNGEIQKLEEKL 3851
> 7300687
Length=4472
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 57/116 (49%), Gaps = 3/116 (2%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKP--VSWEDARKILKATD-FISKILNF 57
K +L ELKS +PP AV VL++ G+ P SW+ A+ + D F+ ++N+
Sbjct 3154 KANLTELKSFGSPPGAVTNVTAAVMVLLSQGGKVPKDRSWKAAKIAMAKVDTFLDSLINY 3213
Query 58 DCTSVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEP 113
D ++ + IQ +++ E + S AA L WV + +KF + VEP
Sbjct 3214 DKENIHPEITKAIQPYLKDPEFEPEFVRSKSGAAAGLCAWVINIIKFYEVYCDVEP 3269
> CE07724
Length=2632
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 78/153 (50%), Gaps = 16/153 (10%)
Query 7 LKSMANPPAMAKVAVEAVAVLITDAGEKPV------------SWEDARKILKATDFISKI 54
LK+M PP ++ +EAV +L+ G KP W +K+L F++KI
Sbjct 2176 LKTMRFPPYAVRLCMEAVCILL---GVKPAKITNEIGEVVNDYWVSGQKLLSDIHFLAKI 2232
Query 55 LNFDCTSVSSATRRRIQTKFLTGD-WDIERINKASRAAGPLAQWVESSVKFVLISEQVEP 113
+F +VS T + I+ K+L+ + +D E + + S AA L +WV + + IS+ VEP
Sbjct 2233 RSFARDTVSKKTVKLIREKYLSKEEFDPENVKQCSLAAEGLCRWVLAIDMYNQISKIVEP 2292
Query 114 LQTEIAQLETEAQENEANLQEQQSLITQLEGKL 146
+ + + E +++ L+ ++ + ++ KL
Sbjct 2293 KRERLRKAEVLVKQHLKQLEVKRKALLKVTEKL 2325
> 7298401
Length=4010
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 34/151 (22%), Positives = 75/151 (49%), Gaps = 11/151 (7%)
Query 7 LKSMANPPAMAKVAVEAVAVL----------ITDAGEKPVSWEDARKILKATDFISKILN 56
+K+M +PP ++ +EAV +L + G W ++++L F+ +LN
Sbjct 2661 VKTMKSPPIGVRIVMEAVCILKDVKPDKVPNPSGLGTVEDYWGPSKRVLSDMKFLDSLLN 2720
Query 57 FDCTSVSSATRRRIQTKFLTGD-WDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQ 115
FD ++ +++ + L+ + +D ++I AS A L +WV + K+ ++++ V P +
Sbjct 2721 FDKDNIPVEVMKKLAQRILSNEAFDPDKIKSASTACEGLCRWVIALTKYDVVAKIVAPKK 2780
Query 116 TEIAQLETEAQENEANLQEQQSLITQLEGKL 146
+A+ E L E+ +++ ++E L
Sbjct 2781 LALAEAEATYNAAMKTLNEKLAMLAKVEANL 2811
> Hs16507235
Length=4523
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 4/126 (3%)
Query 3 SLDELKSMANPPAMAKVAVEAVAVLITDAGEKP--VSWEDARKIL-KATDFISKILNFDC 59
+L ELK+ NPP AV VL+ G P SW+ A+ + K DF+ ++N+D
Sbjct 3208 NLSELKAFPNPPIAVTNVTAAVMVLLAPRGRVPKDRSWKAAKVFMGKVDDFLQALINYDK 3267
Query 60 TSVSSATRRRIQTKFLTG-DWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEI 118
+ + + +L +++ I S AA L WV + +KF + VEP + +
Sbjct 3268 EHIPENCLKVVNEHYLKDPEFNPNLIRTKSFAAAGLCAWVINIIKFYEVYCDVEPKRQAL 3327
Query 119 AQLETE 124
AQ E
Sbjct 3328 AQANLE 3333
> Hs17864092
Length=4024
Score = 50.8 bits (120), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 39/152 (25%), Positives = 78/152 (51%), Gaps = 12/152 (7%)
Query 7 LKSMANPPAMAKVAVEAVAVL----------ITDAGEKPVS-WEDARKILKATDFISKIL 55
+KSM +PPA K+ +EA+ +L T +G+K W A+++L F+ +
Sbjct 2694 VKSMKSPPAGVKLVMEAICILKGIKADKIPDPTGSGKKIEDFWGPAKRLLGDMRFLQSLH 2753
Query 56 NFDCTSVSSATRRRIQTKFLTG-DWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPL 114
+D ++ A I+ ++ D+ E+I AS AA L +WV + + +++ V P
Sbjct 2754 EYDKDNIPPAYMNIIRKNYIPNPDFVPEKIRNASTAAEGLCKWVIAMDSYDKVAKIVAPK 2813
Query 115 QTEIAQLETEAQENEANLQEQQSLITQLEGKL 146
+ ++A E E + L+++Q+ + +++ KL
Sbjct 2814 KIKLAAAEGELKIAMDGLRKKQAALKEVQDKL 2845
> Hs13876382
Length=4486
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 55/116 (47%), Gaps = 3/116 (2%)
Query 1 KKSLDELKSMANPPAMAKVAVEAVAVLITDAGEKP--VSWEDARKILKATD-FISKILNF 57
K +L ELKS +PP AV VL+ G P SW+ A+ + D F+ ++NF
Sbjct 3169 KTNLTELKSFGSPPLAVSNVSAAVMVLMAPRGRVPKDRSWKAAKVTMAKVDGFLDSLINF 3228
Query 58 DCTSVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEP 113
+ ++ + I+ +++ E + S AA L WV + V+F + VEP
Sbjct 3229 NKENIHENCLKAIRPYLQDPEFNPEFVATKSYAAAGLCSWVINIVRFYEVFCDVEP 3284
> Hs14730897
Length=1350
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/127 (23%), Positives = 68/127 (53%), Gaps = 4/127 (3%)
Query 20 AVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCTSVSSATRRRIQTKFLTGDW 79
+EA+++L+ KP W A+++L ++F+ ++L +D ++ ++Q D+
Sbjct 3 VMEAISILLN---AKP-DWPSAKQLLGDSNFLKRLLEYDKENIKPQILAKLQKYINNPDF 58
Query 80 DIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEIAQLETEAQENEANLQEQQSLI 139
E++ K S+A + WV + + + + VEP + ++ + E A L+E+Q+L+
Sbjct 59 VPEKVEKVSKACKSMCMWVRAMDLYSRVVKVVEPKRQKLRAAQAELDITMATLREKQALL 118
Query 140 TQLEGKL 146
Q+E ++
Sbjct 119 RQVEDQI 125
> Hs18604725
Length=538
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 63/123 (51%), Gaps = 5/123 (4%)
Query 3 SLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCTSV 62
SL E++S+ PP + + +E V L+ G SW + L I FD ++
Sbjct 306 SLSEIRSLRMPPDVIRDILEGVLRLM---GIFDTSWVSMKSFLAKRGVREDIATFDARNI 362
Query 63 SSATRRRIQTKFLT--GDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEIAQ 120
S R ++ G +D + +AS AA PLA WV++++++ + E++ PL+TE A
Sbjct 363 SKEIRESVEELLFKNKGSFDPKNAKRASTAAAPLAAWVKANIQYSHVLERIHPLETEQAG 422
Query 121 LET 123
LE+
Sbjct 423 LES 425
> Hs15029526
Length=4490
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 35/150 (23%), Positives = 66/150 (44%), Gaps = 10/150 (6%)
Query 7 LKSMANPPAMAKVAVEAVAVL-------ITDAGEKPV---SWEDARKILKATDFISKILN 56
++ +A PP + ++ V +L +T EK SW ++ K++ AT F+ +
Sbjct 3173 VRKLAKPPHLIMRIMDCVLLLFQKKIDPVTMDPEKSCCKPSWGESLKLMSATGFLWSLQQ 3232
Query 57 FDCTSVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQT 116
F +++ T +Q F D+ E K L W + F I+ +V PL+
Sbjct 3233 FPKDTINEETVELLQPYFNMDDYTFESAKKVCGNVAGLLSWTLAMAIFYGINREVLPLKA 3292
Query 117 EIAQLETEAQENEANLQEQQSLITQLEGKL 146
+A+ E A L + Q+L+ + + +L
Sbjct 3293 NLAKQEGRLAVANAELGKAQALLDEKQAEL 3322
> CE05659
Length=4131
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 64/118 (54%), Gaps = 5/118 (4%)
Query 2 KSLDELKSMANPPAMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCTS 61
+SL E++S+ PP + ++AV + + G SWE RK L + I+NFD
Sbjct 2853 ESLSEIRSLRAPPEAVRDILQAVLLFM---GILDTSWEAMRKFLSKSGVKDDIMNFDANR 2909
Query 62 VSSATRRRIQ--TKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTE 117
+++ +++ K + ++ +AS AA PLA WV++++++ I E++ PL+ E
Sbjct 2910 ITNEIHKKVTALVKQKSNSFEEANAKRASAAAAPLAAWVKANLEYSKILEKIAPLEGE 2967
> Hs22067157
Length=3672
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 29/122 (23%), Positives = 55/122 (45%), Gaps = 18/122 (14%)
Query 7 LKSMANPPAMAKVAVEAVAVLITDAGEKPV--------------SWEDARKILKATDFIS 52
+KSM NPP K+ +E++ ++ G KP W ++KIL F+
Sbjct 2341 VKSMQNPPGPVKLVMESICIM---KGMKPERKPDPSGSGKMIEDYWGVSKKILGDLKFLE 2397
Query 53 KILNFDCTSVSSATRRRIQTKFLTG-DWDIERINKASRAAGPLAQWVESSVKFVLISEQV 111
+ +D ++ T +RI+ +F+ ++ I S A L +WV + + +++ V
Sbjct 2398 SLKTYDKDNIPPLTMKRIRERFINHPEFQPAVIKNVSSACEGLCKWVRAMEVYDRVAKVV 2457
Query 112 EP 113
P
Sbjct 2458 AP 2459
> 7299225
Length=3508
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/143 (20%), Positives = 66/143 (46%), Gaps = 15/143 (10%)
Query 7 LKSMANPPAMAKVAVEAVAVL--------ITDAG---EKPVSWEDARKILKATDFISKIL 55
++ + PP + ++ V +L I DAG KP SW+++ K++ + F+ ++
Sbjct 3296 VRKLGRPPHLIMRIMDCVLILFKRKLHPCIPDAGTPCPKP-SWQESLKMMASATFLLQLQ 3354
Query 56 NFDCTSVSSATRRRIQTKFLTGDWDIERINKASRAAGPLAQWVESSVKFVLISEQVEPLQ 115
N+ +++ +Q F D++++ + L W ++ F ++++V PL+
Sbjct 3355 NYPKDTINDEMIDLLQPYFRMEDYNMDMARRVCGDVAGLLSWTKAMSFFHSVNKEVLPLK 3414
Query 116 TEIAQLETE---AQENEANLQEQ 135
+ E A ++ A +EQ
Sbjct 3415 ANLTMQEARLKLAMDDLAGAEEQ 3437
> Hs19115954
Length=4624
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/117 (21%), Positives = 53/117 (45%), Gaps = 1/117 (0%)
Query 21 VEAVAVLITDAGEKPVSWEDARKILKATDFISKILNFDCTSVSSATRRRIQTKFLTGDWD 80
V AV + + + P SW+++ K++ A +F+ + F +++ + F D++
Sbjct 3334 VSAVKIDLEKSCTMP-SWQESLKLMTAGNFLQNLQQFPKDTINEEVIEFLSPYFEMPDYN 3392
Query 81 IERINKASRAAGPLAQWVESSVKFVLISEQVEPLQTEIAQLETEAQENEANLQEQQS 137
IE + L W ++ F I+++V PL+ + E +LQ+ Q+
Sbjct 3393 IETAKRVCGNVAGLCSWTKAMASFFSINKEVLPLKANLVVQENRHLLAMQDLQKAQA 3449
> 7299152
Length=320
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 30/53 (56%), Gaps = 7/53 (13%)
Query 5 DELKSMANPP--AMAKVAVEAVAVLITDAGEKPVSWEDARKILKATDFISKIL 55
D L MA P M +V+V+ VA GEK V+ E+A K+L A IS++L
Sbjct 177 DHLAEMAKPSNVLMLRVSVDGVA---KAHGEKSVAVEEANKLLSAA--ISRLL 224
> At3g56480
Length=499
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 57/114 (50%), Gaps = 17/114 (14%)
Query 39 EDARKILKATDFISKILNFDCTSVSSATRRRIQTKFLTGDWDIERINKASRAAGP----L 94
EDA+K+ +++ +F C + +A R + + + ERI++ASRA GP L
Sbjct 171 EDAKKL------VNQEKSFSCAEIETA--RAVVLRLGEAFEEQERISEASRAQGPDVEKL 222
Query 95 AQWVESS--VKFVLISEQVEPLQTEIAQLETEAQE---NEANLQEQQSLITQLE 143
+ V+ + +K + +V +Q E+ L QE N L ++ ++I ++E
Sbjct 223 VEEVQEARQIKRMHHPTKVMGMQHELHGLRNRIQEKYMNSVKLHKEIAIIKRVE 276
> CE05294
Length=381
Score = 27.7 bits (60), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 34/65 (52%), Gaps = 7/65 (10%)
Query 33 EKPVSWEDARKILKATDFISKILNFDCTSVSSATRRRIQTKFLTGD------WDIERINK 86
+K VS EDA K+ K + I+K + F+C + ++ + + +G+ W+ + NK
Sbjct 6 QKGVSTEDAVKMTKEQEDIAKYIRFNCPTATTMFEGN-EVHYFSGNKAVDTLWESKYGNK 64
Query 87 ASRAA 91
A + A
Sbjct 65 AKKDA 69
Lambda K H
0.311 0.125 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40