bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5219_orf2
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
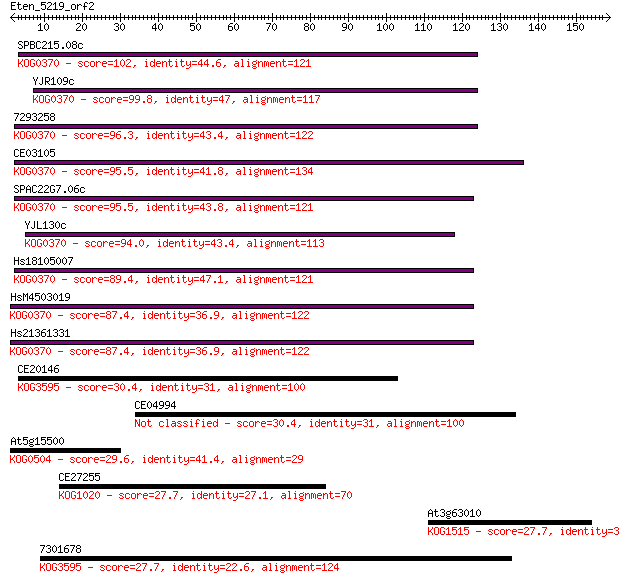
SPBC215.08c 102 4e-22
YJR109c 99.8 2e-21
7293258 96.3 2e-20
CE03105 95.5 4e-20
SPAC22G7.06c 95.5 4e-20
YJL130c 94.0 1e-19
Hs18105007 89.4 3e-18
HsM4503019 87.4 1e-17
Hs21361331 87.4 1e-17
CE20146 30.4 1.4
CE04994 30.4 1.6
At5g15500 29.6 2.7
CE27255 27.7 8.7
At3g63010 27.7 8.8
7301678 27.7 9.2
> SPBC215.08c
Length=1160
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 54/122 (44%), Positives = 77/122 (63%), Gaps = 9/122 (7%)
Query 3 LARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKV-SPAQLTSADLFYVKKYGFSDRQ 61
L +GYS++++H+LT ID WFLSKL ++ ++ + L ++ S L + KK GFSD Q
Sbjct 512 LNQGYSIDKIHDLTKIDKWFLSKLANMAKVYKELEEIGSLYGLNKEIMLRAKKTGFSDLQ 571
Query 62 IASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDD 121
I+ VG +E DV RK L V P++K+IDTLAAEFPA TNYLY +Y+ D
Sbjct 572 ISKLVGA--------SELDVRARRKRLDVHPWVKKIDTLAAEFPAHTNYLYTSYNASSHD 623
Query 122 VE 123
++
Sbjct 624 ID 625
> YJR109c
Length=1118
Score = 99.8 bits (247), Expect = 2e-21, Method: Composition-based stats.
Identities = 55/118 (46%), Positives = 72/118 (61%), Gaps = 3/118 (2%)
Query 7 YSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFY-VKKYGFSDRQIASY 65
Y+VER++EL+ ID WFL K +I + + L V S DL KK GFSD+QIA
Sbjct 460 YTVERVNELSKIDKWFLYKCMNIVNIYKELESVKSLSDLSKDLLQRAKKLGFSDKQIAVT 519
Query 66 VGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDDVE 123
+ + E ++ RK L + P++KRIDTLAAEFPAQTNYLY TY+ +DVE
Sbjct 520 INKHA--STNINELEIRSLRKTLGIIPFVKRIDTLAAEFPAQTNYLYTTYNATKNDVE 575
> 7293258
Length=2189
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 53/122 (43%), Positives = 77/122 (63%), Gaps = 9/122 (7%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDRQ 61
AL G S+ LH+LT+ID WFL KLE I L+ L++ + ++ +A L K++GFSD+Q
Sbjct 831 ALQLGMSLRELHQLTNIDYWFLEKLERIILLQSLLTR-NGSRTDAALLLKAKRFGFSDKQ 889
Query 62 IASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDD 121
IA Y+ + TE V R+ + P++K+IDT+A E+PA TNYLY TY+G D
Sbjct 890 IAKYI--------KSTELAVRHQRQEFGIRPHVKQIDTVAGEWPASTNYLYHTYNGSEHD 941
Query 122 VE 123
V+
Sbjct 942 VD 943
> CE03105
Length=2198
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 56/139 (40%), Positives = 78/139 (56%), Gaps = 14/139 (10%)
Query 2 ALARG-----YSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYG 56
ALARG + VE+ HELT ID WFL ++++I + L K +++ L K+ G
Sbjct 812 ALARGMYYGDFDVEKAHELTRIDRWFLFRMQNIVDIYHRLEKTDVNTVSAELLLEAKQAG 871
Query 57 FSDRQIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYH 116
FSDRQIA +G E V R + P +K+IDT+A E+PAQTNYLY T++
Sbjct 872 FSDRQIAKKIGS--------NEYTVREARFVKGITPCVKQIDTVAGEWPAQTNYLYTTFN 923
Query 117 GVVDDVEPLNTTDLPPVLG 135
G+ +DV N + VLG
Sbjct 924 GIENDVS-FNMKNAVMVLG 941
> SPAC22G7.06c
Length=2244
Score = 95.5 bits (236), Expect = 4e-20, Method: Composition-based stats.
Identities = 53/121 (43%), Positives = 74/121 (61%), Gaps = 8/121 (6%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDRQ 61
A+A GYSV+R+ ELT ID WFL KL + + + +SK + L + L K+ GF+D Q
Sbjct 897 AMASGYSVDRIWELTRIDKWFLEKLMGLIRTSQLISKHDISSLPISLLKTAKQLGFADVQ 956
Query 62 IASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDD 121
IA+++ TE V R R + P++K+IDT+AAEFPA TNYLY TY+ V D
Sbjct 957 IAAFMNS--------TELAVRRIRTEAGIRPFVKQIDTVAAEFPAFTNYLYTTYNAVEHD 1008
Query 122 V 122
+
Sbjct 1009 I 1009
> YJL130c
Length=2214
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 49/114 (42%), Positives = 70/114 (61%), Gaps = 9/114 (7%)
Query 5 RGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKV-SPAQLTSADLFYVKKYGFSDRQIA 63
+GYSV+++ E+T ID WFL+KL + Q E +S + +L S L K+ GF DRQIA
Sbjct 865 KGYSVDKVWEMTRIDKWFLNKLHDLVQFAEKISSFGTKEELPSLVLRQAKQLGFDDRQIA 924
Query 64 SYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHG 117
++ V + R RK + P++K+IDT+AAEFPA TNYLY+TY+
Sbjct 925 RFLDSNEVA--------IRRLRKEYGITPFVKQIDTVAAEFPAYTNYLYMTYNA 970
> Hs18105007
Length=2225
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 57/125 (45%), Positives = 74/125 (59%), Gaps = 16/125 (12%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLE----HIQQLKESLSKVSPAQLTSADLFYVKKYGF 57
AL GYSV+RL+ELT ID WFL +++ H Q L++ + P L L K GF
Sbjct 816 ALWAGYSVDRLYELTRIDRWFLHRMKRIIAHAQLLEQHRGQPLPPDL----LQQAKCLGF 871
Query 58 SDRQIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHG 117
SD+QIA V TE V + R+ L + P +K+IDT+AAE+PAQTNYLYLTY G
Sbjct 872 SDKQIALAV--------LSTELAVRKLRQELGICPAVKQIDTVAAEWPAQTNYLYLTYWG 923
Query 118 VVDDV 122
D+
Sbjct 924 TTHDL 928
> HsM4503019
Length=1500
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 45/122 (36%), Positives = 73/122 (59%), Gaps = 8/122 (6%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+A+ S++ + +LT+ID WFL K+ I ++++L ++ +T L K+ GFSD+
Sbjct 856 KAIDDNMSLDEIEKLTYIDKWFLYKMRDILNMEKTLKGLNSESMTEETLKRAKEIGFSDK 915
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
QI+ +G +TE R + P++K+IDTLAAE+P+ TNYLY+TY+G
Sbjct 916 QISKCLG--------LTEAQTRELRLKKNIHPWVKQIDTLAAEYPSVTNYLYVTYNGQEH 967
Query 121 DV 122
DV
Sbjct 968 DV 969
> Hs21361331
Length=1500
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 45/122 (36%), Positives = 73/122 (59%), Gaps = 8/122 (6%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+A+ S++ + +LT+ID WFL K+ I ++++L ++ +T L K+ GFSD+
Sbjct 856 KAIDDNMSLDEIEKLTYIDKWFLYKMRDILNMEKTLKGLNSESMTEETLKRAKEIGFSDK 915
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
QI+ +G +TE R + P++K+IDTLAAE+P+ TNYLY+TY+G
Sbjct 916 QISKCLG--------LTEAQTRELRLKKNIHPWVKQIDTLAAEYPSVTNYLYVTYNGQEH 967
Query 121 DV 122
DV
Sbjct 968 DV 969
> CE20146
Length=2265
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 31/117 (26%), Positives = 48/117 (41%), Gaps = 17/117 (14%)
Query 3 LARGYSVERL---HELTHIDPWFLSKLEHIQQLKESLSKVSPAQL-------TSADLFYV 52
L +VE L HE + P F + + ++ S PAQL +S +
Sbjct 1242 LPTNIAVEHLGVFHEFVEVSPAFKFETSPLSSVRTSSIPKLPAQLVSTGQPPSSPHSVLL 1301
Query 53 KKYG--FSDRQIASYVGGGPVEGLRVT-----EEDVWRYRKALVVEPYIKRIDTLAA 102
+K G +R+I+ VG E L+ T E+ +W Y V + + R D AA
Sbjct 1302 EKIGAKIRNREISGCVGSLAKETLKETLQNFPEKIIWVYTDVFVRDQMVTRSDEFAA 1358
> CE04994
Length=405
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 49/115 (42%), Gaps = 19/115 (16%)
Query 34 ESLSKVSPAQLTSADLF--YVKKYGFSDRQI----------ASYVGGG---PVEGLRVTE 78
E+ S V P ++T+A F + + DR+ AS V G P E R
Sbjct 288 EAASSVHPPKITTASKFDRQTDRRSYGDRRYLFENPEEEKQASLVRGTGRPPAEWGRKQN 347
Query 79 EDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDDVEPLNTTDLPPV 133
E++ + RK L Y++++ A P + L++ DD EP T+ P V
Sbjct 348 EELEQIRKNLEPPKYVEQVKVEGARAPVEPVPLFVP----TDDFEPQQVTEDPNV 398
> At5g15500
Length=457
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHI 29
EA A+ +++ L+EL H DP+ L K +H+
Sbjct 7 EAAAKSGNIDLLYELIHEDPYVLDKTDHV 35
> CE27255
Length=2227
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 19/74 (25%), Positives = 36/74 (48%), Gaps = 7/74 (9%)
Query 14 ELTHIDPWFLSKLEHIQQLKES----LSKVSPAQLTSADLFYVKKYGFSDRQIASYVGGG 69
++ +D +S+L + Q++ L P+ L F ++ YGF++ ++A Y
Sbjct 2074 DIVFLDENMMSRLSQLGQIETFHQLFLDSQVPSLLLYVRTFLMQLYGFNETKVAEY---Q 2130
Query 70 PVEGLRVTEEDVWR 83
P E +V E+ V R
Sbjct 2131 PSEAAKVYEKAVTR 2144
> At3g63010
Length=358
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 25/43 (58%), Gaps = 5/43 (11%)
Query 111 LYLTYHGVVDDVEPLNTTDLPPVLGPWGGGHQESAQFTAPSAQ 153
L+ T HG ++ +PL+TT++ PVL + GG FT SA
Sbjct 86 LHQTRHGTLELTKPLSTTEIVPVLIFFHGG-----SFTHSSAN 123
> 7301678
Length=4820
Score = 27.7 bits (60), Expect = 9.2, Method: Composition-based stats.
Identities = 28/136 (20%), Positives = 59/136 (43%), Gaps = 14/136 (10%)
Query 9 VERLHE-LTHIDPWFLSKLEHIQQLKESLSKVSP---------AQLTSADLFYVKKYGFS 58
+++L E L ++ + + ++ ++ L E + + + LTS + + K+
Sbjct 3844 IQKLEEKLNELNENYATSMKQMRALTEMMQQAERRLIASDKLISGLTSELIRWSKEMASL 3903
Query 59 DRQIASYVGGGPVEG--LRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYH 116
+Q+ VGG + L T W +RKA+V + +++ I +L P Q + Y
Sbjct 3904 GQQLIDSVGGCLISASFLAYTGAFTWEFRKAMVFDDWLEDIASLG--IPIQLPFKIDGYL 3961
Query 117 GVVDDVEPLNTTDLPP 132
++ + LPP
Sbjct 3962 TTDVEISQWSNEGLPP 3977
Lambda K H
0.316 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2100092188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40