bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5261_orf2
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
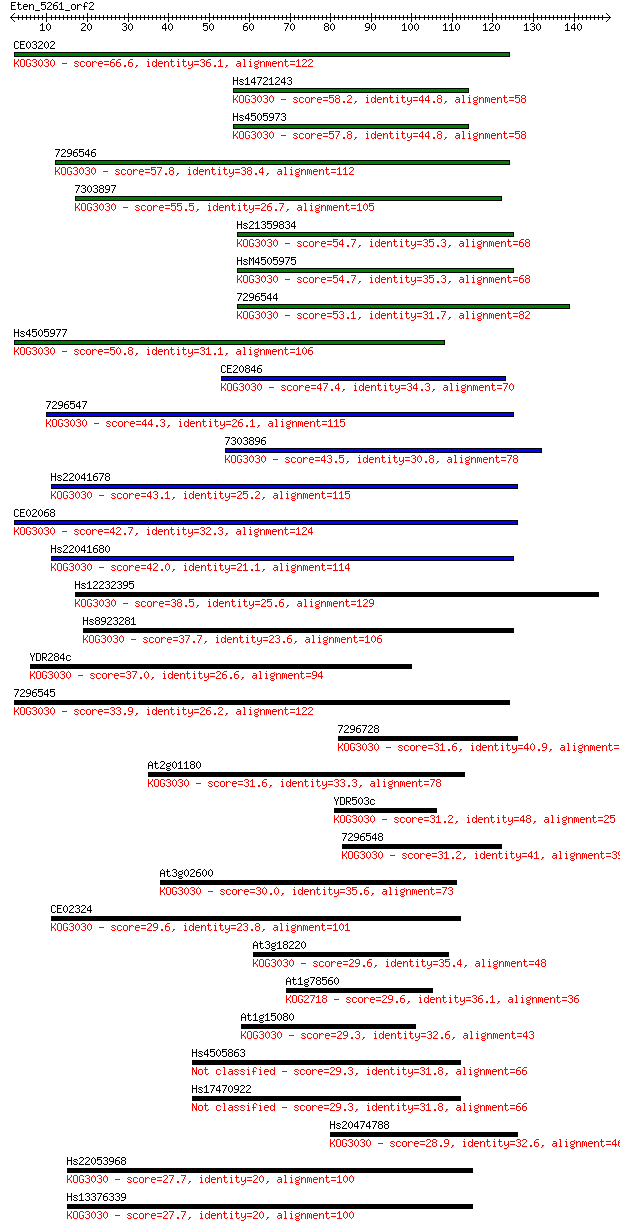
CE03202 66.6 2e-11
Hs14721243 58.2 5e-09
Hs4505973 57.8 7e-09
7296546 57.8 8e-09
7303897 55.5 3e-08
Hs21359834 54.7 6e-08
HsM4505975 54.7 6e-08
7296544 53.1 2e-07
Hs4505977 50.8 9e-07
CE20846 47.4 1e-05
7296547 44.3 8e-05
7303896 43.5 1e-04
Hs22041678 43.1 2e-04
CE02068 42.7 2e-04
Hs22041680 42.0 4e-04
Hs12232395 38.5 0.004
Hs8923281 37.7 0.009
YDR284c 37.0 0.012
7296545 33.9 0.12
7296728 31.6 0.52
At2g01180 31.6 0.64
YDR503c 31.2 0.66
7296548 31.2 0.76
At3g02600 30.0 1.5
CE02324 29.6 2.1
At3g18220 29.6 2.2
At1g78560 29.6 2.5
At1g15080 29.3 2.6
Hs4505863 29.3 3.1
Hs17470922 29.3 3.1
Hs20474788 28.9 3.4
Hs22053968 27.7 7.3
Hs13376339 27.7 7.3
> CE03202
Length=318
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 44/123 (35%), Positives = 63/123 (51%), Gaps = 17/123 (13%)
Query 2 RKGSINLGQLMTLSFVAPSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTVPQYIVDM 61
++ ++ L L+ ++ +P LI+ LVE ++ ++ A+ F T Y+
Sbjct 94 KENTVGLKHLLVITLGSPFLIVALVEAILHFKSKGSNRLAKF------FSATTITYL--- 144
Query 62 YKYLGGFGFTMATAWLFA-DSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYVEDFH 120
KYL M A FA + LKC+VG LRPHFF CKPDWSKV C + +I D
Sbjct 145 -KYL-----LMYAACTFAMEFLKCYVGRLRPHFFSVCKPDWSKVDCTDKQS-FIDSSDLV 197
Query 121 CTN 123
CTN
Sbjct 198 CTN 200
> Hs14721243
Length=285
Score = 58.2 bits (139), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 56 QYIVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEY 113
YI +YK +G F F A + D K +G LRPHF D C PDWSK+ C EY
Sbjct 94 NYIATIYKAIGTFLFGAAASQSLTDIAKYSIGRLRPHFLDVCDPDWSKINCSDGYIEY 151
> Hs4505973
Length=289
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 56 QYIVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEY 113
YI +YK +G F F A + D K +G LRPHF D C PDWSK+ C EY
Sbjct 98 NYIATIYKAIGTFLFGAAASQSLTDIAKYSIGRLRPHFLDVCDPDWSKINCSDGYIEY 155
> 7296546
Length=305
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 62/125 (49%), Gaps = 23/125 (18%)
Query 12 MTLSFVA-PSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTVPQYIVDMYK----YLG 66
+T++ VA P+ +++VE+L A V V E + G +P++I + YK YL
Sbjct 63 LTIAVVALPAAFVLVVEMLRAAV-VPSSTE--LTQRFVFVGVRIPRFISECYKAIGVYLF 119
Query 67 GFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDW--------SKVTCKGENGEYIYVED 118
G G T+A L S G LRP+FFD C+P W S VT + +Y+ED
Sbjct 120 GLGLTLAAIRLTKHS----TGRLRPYFFDICQPTWGTEGGESCSDVTAQNST---LYLED 172
Query 119 FHCTN 123
F CT
Sbjct 173 FSCTE 177
> 7303897
Length=372
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 50/106 (47%), Gaps = 1/106 (0%)
Query 17 VAPSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTVPQYIVDMYKYLGGFGFTMATAW 76
V P +I +VE++I+ + +D + +P ++++ YK +G + F +
Sbjct 138 VIPVGVIFIVEVIISQNKAKQDNGNATSRRYVFMNYELPDWMIECYKKIGIYAFGAVLSQ 197
Query 77 LFADSLKCFVGSLRPHFFDACKPDWSK-VTCKGENGEYIYVEDFHC 121
L D K +G LRPHF C+P + TC Y+++F C
Sbjct 198 LTTDIAKYSIGRLRPHFIAVCQPQMADGSTCDDAINAGKYIQEFTC 243
> Hs21359834
Length=311
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 5/68 (7%)
Query 57 YIVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYV 116
Y+ +YK +G F F A + F D K +G LRPHF C PD+S++ C Y+
Sbjct 122 YVAALYKQVGCFLFGCAISQSFTDIAKVSIGRLRPHFLSVCNPDFSQINCSEG-----YI 176
Query 117 EDFHCTND 124
+++ C D
Sbjct 177 QNYRCRGD 184
> HsM4505975
Length=311
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 5/68 (7%)
Query 57 YIVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYV 116
Y+ +YK +G F F A + F D K +G LRPHF C PD+S++ C Y+
Sbjct 122 YVAALYKQVGCFLFGCAISQSFTDIAKVSIGRLRPHFLSVCNPDFSQINCSEG-----YI 176
Query 117 EDFHCTND 124
+++ C D
Sbjct 177 QNYRCRGD 184
> 7296544
Length=334
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 40/83 (48%), Gaps = 1/83 (1%)
Query 57 YIVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSK-VTCKGENGEYIY 115
Y ++++ F F +L + K VG LRPHFF C+P +C +Y
Sbjct 58 YFRNLWRAEATFSFGFIATYLTTELAKHAVGRLRPHFFHGCQPRLDDGSSCSDLQNAELY 117
Query 116 VEDFHCTNDPLFSSRLCSVFCQF 138
VE FHCTN+ L + ++ + F
Sbjct 118 VEQFHCTNNNLSTRQIRELHVSF 140
> Hs4505977
Length=288
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 47/106 (44%), Gaps = 12/106 (11%)
Query 2 RKGSINLGQLMTLSFVAPSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTVPQYIVDM 61
R +I G + ++ A +ILV A + T+ +R N Y+ +
Sbjct 48 RPDTITHGLMAGVTITA---TVILVSAGEAYLVYTDRLYSRSDFN---------NYVAAV 95
Query 62 YKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCK 107
YK LG F F A + D K +G LRP+F C PDWS+V C
Sbjct 96 YKVLGTFLFGAAVSQSLTDLAKYMIGRLRPNFLAVCDPDWSRVNCS 141
> CE20846
Length=346
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 36/70 (51%), Gaps = 3/70 (4%)
Query 53 TVPQYIVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGE 112
+V + +V +Y ++G F + L D K +G RPHF D C+PD TC +
Sbjct 101 SVHRLVVRLYCFIGYFFVGVCFNQLMVDIAKYTIGRQRPHFMDVCRPDIGYQTCSQPD-- 158
Query 113 YIYVEDFHCT 122
+Y+ DF CT
Sbjct 159 -LYITDFKCT 167
> 7296547
Length=340
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 56/118 (47%), Gaps = 3/118 (2%)
Query 10 QLMTLSFVAPSLIIILVELLIAVV--RVTEDKEARMKSNVPMFGWTVPQYIVDMYKYLGG 67
Q T++ V LI+ L+ L+ VV V+ + + + V + GW V + V++ +
Sbjct 78 QDNTITPVMLGLIVGLLPALVMVVVEYVSHLRAGDISATVDLLGWRVSTWYVELGRQSTY 137
Query 68 FGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVT-CKGENGEYIYVEDFHCTND 124
F F + + + K +G LRPHF C+P + + C + Y+E++ C +
Sbjct 138 FCFGLLLTFDATEVGKYTIGRLRPHFLAVCQPQIADGSMCSDPVNLHRYMENYDCAGE 195
> 7303896
Length=246
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 38/79 (48%), Gaps = 1/79 (1%)
Query 54 VPQYIVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSK-VTCKGENGE 112
+P ++V+ Y +G F F + L + K +G LRPHF+ C+P TC
Sbjct 81 LPTWLVECYHRMGIFIFGLGVEQLSTNIAKYSIGRLRPHFYTLCQPVMKDGTTCSDPINA 140
Query 113 YIYVEDFHCTNDPLFSSRL 131
Y+E+F C + S +L
Sbjct 141 ARYIEEFTCAAVDITSKQL 159
> Hs22041678
Length=321
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 61/116 (52%), Gaps = 3/116 (2%)
Query 11 LMTLSFVAPSLIIILVELLIAVVRV-TEDKEARMKSNVPMFGWTVPQYIVDMYKYLGGFG 69
L +L+ P L+II+ E + +++ T D E + K+ + + + ++LG +
Sbjct 68 LYSLAAGVPVLVIIVGETAVFCLQLATRDFENQEKTILTGDCCYINPLVRRTVRFLGIYT 127
Query 70 FTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYVEDFHCTNDP 125
F + +F ++ + G+L PHF CKP+++ + C+ + ++I E+ CT +P
Sbjct 128 FGLFATDIFVNAGQVVTGNLAPHFLALCKPNYTALGCQ-QYTQFISGEE-ACTGNP 181
> CE02068
Length=341
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 56/132 (42%), Gaps = 23/132 (17%)
Query 2 RKGSINLGQLMTLSFVAPSLIIILVELLIAVVRVTEDKEARMKSNV--PMFGWTVPQY-- 57
RK +I QLM + V + ++ VE + +++SN+ P + W
Sbjct 67 RKDTITAVQLMLYNLVLNAATVLFVEYY---------RMQKVESNINNPRYRWRNNHLHV 117
Query 58 -IVDMYKYLG--GFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTC-KGENGEY 113
V + Y G GF M A K VG LRPHF D CK + TC G++ Y
Sbjct 118 LFVRLLTYFGYSQIGFVMNIALNIVT--KHVVGRLRPHFLDVCK--LANDTCVTGDSHRY 173
Query 114 IYVEDFHCTNDP 125
I D+ CT P
Sbjct 174 I--TDYTCTGPP 183
> Hs22041680
Length=763
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 24/114 (21%), Positives = 52/114 (45%), Gaps = 1/114 (0%)
Query 11 LMTLSFVAPSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTVPQYIVDMYKYLGGFGF 70
L++L+F P++ I++ E ++ ++ N+ G ++ +++G F
Sbjct 125 LLSLAFAGPAITIMVGEGILYCCLSKRRNGVGLEPNINAGGCNFNSFLRRAVRFVGVHVF 184
Query 71 TMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYVEDFHCTND 124
+ + L D ++ G P+F CKP+++ + + YI VED +D
Sbjct 185 GLCSTALITDIIQLSTGYQAPYFLTVCKPNYTSLNVSCKENSYI-VEDICSGSD 237
> Hs12232395
Length=343
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/144 (22%), Positives = 57/144 (39%), Gaps = 22/144 (15%)
Query 17 VAPSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTV---------PQYIVDMYKYLGG 67
P+L I+L EL A S VP+ G + + + ++LG
Sbjct 80 AGPTLTILLGELARAFFPAPP-------SAVPVIGESTIVSGACCRFSPPVRRLVRFLGV 132
Query 68 FGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTC----KGENGEYIYVED-FHCT 122
+ F + T +FA++ + G+ PHF C+P+++ + C G +V D C
Sbjct 133 YSFGLFTTTIFANAGQVVTGNPTPHFLSVCRPNYTALGCLPPSPDRPGPDRFVTDQGACA 192
Query 123 NDPLFSSRLCSVF-CQFRLVCTYV 145
P + F C+ +C Y
Sbjct 193 GSPSLVAAARRAFPCKDAALCAYA 216
> Hs8923281
Length=325
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/107 (23%), Positives = 52/107 (48%), Gaps = 3/107 (2%)
Query 19 PSLIIILVELLIAVVRVT-EDKEARMKSNVPMFGWTVPQYIVDMYKYLGGFGFTMATAWL 77
P+ II + E+ + ++ T E AR K+ + + + + ++ G F F + +
Sbjct 81 PTAIIFIGEISMYFIKSTRESLIAREKTILTGECCYLNPLLRRIIRFTGVFAFGLFATDI 140
Query 78 FADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYVEDFHCTND 124
F ++ + G L P+F CKP+++ C+ + ++I + CT D
Sbjct 141 FVNAGQVVTGHLTPYFLTVCKPNYTSADCQAHH-QFINNGNI-CTGD 185
> YDR284c
Length=289
Score = 37.0 bits (84), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 38/94 (40%), Gaps = 21/94 (22%)
Query 6 INLGQLMTLSFVAPSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTVPQYIVDMYKYL 65
+N L SFV PSL I+++ ++A R I +Y L
Sbjct 62 VNNNMLFVYSFVVPSLTILIIGSILADRR---------------------HLIFILYTSL 100
Query 66 GGFGFTMATAWLFADSLKCFVGSLRPHFFDACKP 99
G + F + +K ++G LRP F D C+P
Sbjct 101 LGLSLAWFSTSFFTNFIKNWIGRLRPDFLDRCQP 134
> 7296545
Length=341
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 53/125 (42%), Gaps = 10/125 (8%)
Query 2 RKGSINLGQLMTLSFVAPSLIIILVELLIAVVRVTEDKEARMKSNVPMFGWTVPQYIVDM 61
R G+I+ ++ + P+ +I++VEL + +EA K + + +
Sbjct 50 RDGTISSKVIIAIVLGVPNAVIVVVELFRQLPG-GPLREAGGKRDSCRIAHRLGVLYRQV 108
Query 62 YKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACK---PDWSKVTCKGENGEYIYVED 118
YL G T L K +G LRPHF C+ PD S +C+ Y++
Sbjct 109 IFYLYGLAMVTFTTML----TKLCLGRLRPHFLAVCQPMLPDGS--SCQDAQNLGRYIDS 162
Query 119 FHCTN 123
F C+N
Sbjct 163 FTCSN 167
> 7296728
Length=412
Score = 31.6 bits (70), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 82 LKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYVEDFHCTNDP 125
LK VG RP +F C PD V NG + DF+CT P
Sbjct 215 LKITVGRPRPDYFYRCFPDGVMVLNTTSNGVDTSILDFNCTGLP 258
> At2g01180
Length=302
Score = 31.6 bits (70), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 26/103 (25%), Positives = 41/103 (39%), Gaps = 27/103 (26%)
Query 35 VTEDKEARMKSNVPMFGWTVPQYIV------------------DMYKYLGGFGFTMATAW 76
+T+ K + VP+ W+VP Y V D++ + G F +
Sbjct 54 MTDLKYPFKDNTVPI--WSVPVYAVLLPIIVFVCFYLKRTCVYDLHHSILGLLFAVLITG 111
Query 77 LFADSLKCFVGSLRPHFFDACKPDWSK-------VTCKGENGE 112
+ DS+K G RP+F+ C PD + V C G+ E
Sbjct 112 VITDSIKVATGRPRPNFYWRCFPDGKELYDALGGVVCHGKAAE 154
> YDR503c
Length=274
Score = 31.2 bits (69), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 81 SLKCFVGSLRPHFFDACKPDWSKVT 105
+LK +G+LRP F D C PD K++
Sbjct 134 ALKLIIGNLRPDFVDRCIPDLQKMS 158
> 7296548
Length=305
Score = 31.2 bits (69), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 19/42 (45%), Gaps = 3/42 (7%)
Query 83 KCFVGSLRPHFFDACK---PDWSKVTCKGENGEYIYVEDFHC 121
K +G LRPHFF C PD S + G Y D+ C
Sbjct 119 KQALGRLRPHFFAVCSPHFPDGSSCLDESHRGALKYHTDYEC 160
> At3g02600
Length=314
Score = 30.0 bits (66), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 40/99 (40%), Gaps = 28/99 (28%)
Query 38 DKEARMKSN-VPMFGWTVPQY------------------IVDMYKYLGGFGFTMATAWLF 78
D +KSN VP+ W+VP Y + D++ + G +++ +
Sbjct 56 DLSYPLKSNTVPI--WSVPVYAMLLPLVIFIFIYFRRRDVYDLHHAVLGLLYSVLVTAVL 113
Query 79 ADSLKCFVGSLRPHFFDACKPD-------WSKVTCKGEN 110
D++K VG RP FF C PD V C G+
Sbjct 114 TDAIKNAVGRPRPDFFWRCFPDGKALYDSLGDVICHGDK 152
> CE02324
Length=396
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 24/104 (23%), Positives = 44/104 (42%), Gaps = 10/104 (9%)
Query 11 LMTLSFVAPSLIIILVELLIAVVRVTEDKEARMKSN---VPMFGWTVPQYIVDMYKYLGG 67
L TL+F P L+I++ E++ + K V +F + ++++ YL G
Sbjct 97 LYTLAFTIPPLVILIGEVMFWLFSTKPRKIVYANCGECPVHLFTRRLFRFVI---IYLAG 153
Query 68 FGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENG 111
+ +F D++K G RP+F C + T E+
Sbjct 154 ----LLIVQIFVDTIKLMTGYQRPYFLSLCNVSITACTAPLEHS 193
> At3g18220
Length=301
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 61 MYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWS-KVTCKG 108
++ + G GF+ + DS+K VG RP+FF C P+ V C G
Sbjct 96 LHHAILGIGFSCLVTGVTTDSIKDAVGRPRPNFFYRCFPNGKPDVVCHG 144
> At1g78560
Length=401
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 69 GFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSKV 104
G ++TA+ SL C +G +RP F+ P+W+ V
Sbjct 94 GEAVSTAFPIWVSLGCLLGLMRPSTFNWVTPNWTIV 129
> At1g15080
Length=290
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 58 IVDMYKYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPD 100
+ D++ + G F++ + D++K VG RP FF C PD
Sbjct 93 VYDLHHAILGLLFSVLITGVITDAIKDAVGRPRPDFFWRCFPD 135
> Hs4505863
Length=431
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 34/82 (41%), Gaps = 25/82 (30%)
Query 46 NVPMFGWTVPQYIVDMYKYLGGFGFTMATAWLFADSLKCFVGSL-------RPHFFD--- 95
N P FG + + GFG +T +L+ + LK V L +PH++
Sbjct 305 NDPQFGTSCE---------ITGFGKENSTDYLYPEQLKMTVVKLISHRECQQPHYYGSEV 355
Query 96 ------ACKPDWSKVTCKGENG 111
A P W +C+G++G
Sbjct 356 TTKMLCAADPQWKTDSCQGDSG 377
> Hs17470922
Length=431
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 34/82 (41%), Gaps = 25/82 (30%)
Query 46 NVPMFGWTVPQYIVDMYKYLGGFGFTMATAWLFADSLKCFVGSL-------RPHFFD--- 95
N P FG + + GFG +T +L+ + LK V L +PH++
Sbjct 305 NDPQFGTSCE---------ITGFGKENSTDYLYPEQLKMTVVKLISHRECQQPHYYGSEV 355
Query 96 ------ACKPDWSKVTCKGENG 111
A P W +C+G++G
Sbjct 356 TTKMLCAADPQWKTDSCQGDSG 377
> Hs20474788
Length=271
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 20/46 (43%), Gaps = 12/46 (26%)
Query 80 DSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYIYVEDFHCTNDP 125
+++K VG RP FF C PD + + HCT DP
Sbjct 99 NTIKLIVGRPRPDFFYRCFPDG------------VMNSEMHCTGDP 132
> Hs22053968
Length=686
Score = 27.7 bits (60), Expect = 7.3, Method: Composition-based stats.
Identities = 20/104 (19%), Positives = 43/104 (41%), Gaps = 4/104 (3%)
Query 15 SFVAPSLIIILVELLIAVVRVT----EDKEARMKSNVPMFGWTVPQYIVDMYKYLGGFGF 70
+F AP+ I++ E ++ ++ A + ++ G ++ +++G F
Sbjct 20 AFAAPAASIMVAEGMLYCLQSRLWGRAGGPAGAEGSINAGGCNFNSFLRRTVRFVGVHVF 79
Query 71 TMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYI 114
+ L D ++ G P F CKP+++ + E YI
Sbjct 80 GLCATALVTDVIQLATGYHTPFFLTVCKPNYTLLGTSCEVNPYI 123
> Hs13376339
Length=746
Score = 27.7 bits (60), Expect = 7.3, Method: Composition-based stats.
Identities = 20/104 (19%), Positives = 43/104 (41%), Gaps = 4/104 (3%)
Query 15 SFVAPSLIIILVELLIAVVRVT----EDKEARMKSNVPMFGWTVPQYIVDMYKYLGGFGF 70
+F AP+ I++ E ++ ++ A + ++ G ++ +++G F
Sbjct 80 AFAAPAASIMVAEGMLYCLQSRLWGRAGGPAGAEGSINAGGCNFNSFLRRTVRFVGVHVF 139
Query 71 TMATAWLFADSLKCFVGSLRPHFFDACKPDWSKVTCKGENGEYI 114
+ L D ++ G P F CKP+++ + E YI
Sbjct 140 GLCATALVTDVIQLATGYHTPFFLTVCKPNYTLLGTSCEVNPYI 183
Lambda K H
0.328 0.141 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40