bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
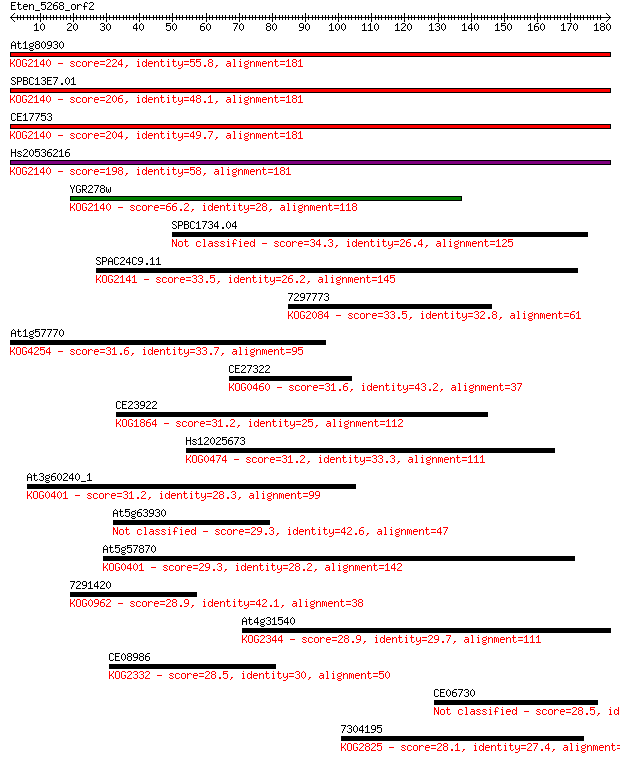
Query= Eten_5268_orf2
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
At1g80930 224 7e-59
SPBC13E7.01 206 2e-53
CE17753 204 9e-53
Hs20536216 198 5e-51
YGR278w 66.2 4e-11
SPBC1734.04 34.3 0.15
SPAC24C9.11 33.5 0.24
7297773 33.5 0.25
At1g57770 31.6 0.83
CE27322 31.6 0.88
CE23922 31.2 1.0
Hs12025673 31.2 1.1
At3g60240_1 31.2 1.2
At5g63930 29.3 4.5
At5g57870 29.3 5.0
7291420 28.9 6.2
At4g31540 28.9 6.4
CE08986 28.5 6.8
CE06730 28.5 7.2
7304195 28.1 9.6
> At1g80930
Length=900
Score = 224 bits (571), Expect = 7e-59, Method: Composition-based stats.
Identities = 101/181 (55%), Positives = 139/181 (76%), Gaps = 0/181 (0%)
Query 1 PFKLARLQQQTSDSKSHAYQRQMWEALRKSINGLVNKANVSNLTQLVQELFRENLVRGRG 60
PFKLAR+ ++ D S YQR W+ALRKSINGLVNK N SN+ ++ ELF ENL+RGRG
Sbjct 336 PFKLARMMKEVEDKSSVEYQRLTWDALRKSINGLVNKVNASNIKNIIPELFAENLIRGRG 395
Query 61 LFCRALIRSQLASPGFTGVYASLLCIVNSKLPDIGELVLKRLILQFRRAYRRNDKVVCLA 120
LFCR+ ++SQ+ASPGFT V+A+L+ ++N+K P++ EL+LKR++LQ +RAY+RNDK LA
Sbjct 396 LFCRSCMKSQMASPGFTDVFAALVAVINAKFPEVAELLLKRVVLQLKRAYKRNDKPQLLA 455
Query 121 CVEFFAHLVNQRIAHELLALQLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAA 180
V+F AHLVNQ++A E++AL+L +LL PT+DSVEV +GF+ G +L+DV G +
Sbjct 456 AVKFIAHLVNQQVAEEIIALELVTILLGDPTDDSVEVAVGFVTECGAMLQDVSPRGLNGI 515
Query 181 F 181
F
Sbjct 516 F 516
> SPBC13E7.01
Length=628
Score = 206 bits (524), Expect = 2e-53, Method: Composition-based stats.
Identities = 87/181 (48%), Positives = 134/181 (74%), Gaps = 0/181 (0%)
Query 1 PFKLARLQQQTSDSKSHAYQRQMWEALRKSINGLVNKANVSNLTQLVQELFRENLVRGRG 60
P KL LQ Q +D + YQR WEAL+KSINGL+NK N SN+ ++ ELF+EN++RGR
Sbjct 97 PAKLKALQAQLTDVNTPEYQRMQWEALKKSINGLINKVNKSNIRDIIPELFQENIIRGRA 156
Query 61 LFCRALIRSQLASPGFTGVYASLLCIVNSKLPDIGELVLKRLILQFRRAYRRNDKVVCLA 120
L+CR+++++Q AS FT +YA++ ++N+K P IGEL+L RLI+QFR++++RNDK +C++
Sbjct 157 LYCRSIMKAQAASLPFTPIYAAMTAVINTKFPQIGELLLTRLIVQFRKSFQRNDKSMCIS 216
Query 121 CVEFFAHLVNQRIAHELLALQLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAA 180
F AHL+NQ+IAHE++ LQ+ +LL++PTNDS+E+ + L+ +G L +V ++
Sbjct 217 SSSFIAHLINQKIAHEIVGLQILAVLLERPTNDSIEIAVMLLREIGAYLAEVSTRAYNGV 276
Query 181 F 181
F
Sbjct 277 F 277
> CE17753
Length=897
Score = 204 bits (518), Expect = 9e-53, Method: Composition-based stats.
Identities = 90/181 (49%), Positives = 135/181 (74%), Gaps = 0/181 (0%)
Query 1 PFKLARLQQQTSDSKSHAYQRQMWEALRKSINGLVNKANVSNLTQLVQELFRENLVRGRG 60
P KL +QQQ SD +S YQR WE ++K I+GLVN+ N NL Q+V+EL +EN++R +G
Sbjct 167 PAKLRLMQQQISDKQSEQYQRMNWERMKKKIHGLVNRVNAKNLVQIVRELLQENVIRSKG 226
Query 61 LFCRALIRSQLASPGFTGVYASLLCIVNSKLPDIGELVLKRLILQFRRAYRRNDKVVCLA 120
L CR +I++Q SPGF+ VYA+L ++NSK P +GEL+L+RLI+QF+R++RRND+ V +
Sbjct 227 LLCRDIIQAQAFSPGFSNVYAALAAVINSKFPHVGELLLRRLIVQFKRSFRRNDRGVTVN 286
Query 121 CVEFFAHLVNQRIAHELLALQLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAA 180
++F AHL+NQ++AHE+LAL++ L+L++PT+DSVEV I FLK G L ++ ++
Sbjct 287 VIKFIAHLINQQVAHEVLALEIMILMLEEPTDDSVEVAIAFLKECGAKLLEIAPAALNSV 346
Query 181 F 181
+
Sbjct 347 Y 347
> Hs20536216
Length=908
Score = 198 bits (503), Expect = 5e-51, Method: Composition-based stats.
Identities = 105/181 (58%), Positives = 138/181 (76%), Gaps = 0/181 (0%)
Query 1 PFKLARLQQQTSDSKSHAYQRQMWEALRKSINGLVNKANVSNLTQLVQELFRENLVRGRG 60
P KL +Q+Q +D S AYQR WEAL+KSINGL+NK N+SN++ ++QEL +EN+VRGRG
Sbjct 136 PAKLRMMQEQITDKNSLAYQRMSWEALKKSINGLINKVNISNISIIIQELLQENIVRGRG 195
Query 61 LFCRALIRSQLASPGFTGVYASLLCIVNSKLPDIGELVLKRLILQFRRAYRRNDKVVCLA 120
L R+++++Q ASP FT VYA+L+ I+NSK P IGEL+LKRLIL FR+ YRRNDK +CL
Sbjct 196 LLSRSVLQAQSASPIFTHVYAALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLT 255
Query 121 CVEFFAHLVNQRIAHELLALQLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAA 180
+F AHL+NQ +AHE+L L++ LLL++PT+DSVEV IGFLK G L V G +A
Sbjct 256 ASKFVAHLINQNVAHEVLCLEMLTLLLERPTDDSVEVAIGFLKECGLKLTQVSPRGINAI 315
Query 181 F 181
F
Sbjct 316 F 316
> YGR278w
Length=577
Score = 66.2 bits (160), Expect = 4e-11, Method: Composition-based stats.
Identities = 33/120 (27%), Positives = 66/120 (55%), Gaps = 2/120 (1%)
Query 19 YQRQMWEALRKSINGLVNKANVSNLTQLVQELFRENLVRGRGLFCRALIRSQLASPG--F 76
+QR+ WE +R ++ +++ + NL + ++LF+ N++ GR + C+ ++ L
Sbjct 13 FQRENWEMIRSHVSPIISNLTMDNLQESHRDLFQVNILIGRNIICKNVVDFTLNKQNGRL 72
Query 77 TGVYASLLCIVNSKLPDIGELVLKRLILQFRRAYRRNDKVVCLACVEFFAHLVNQRIAHE 136
++L+ ++NS +PDIGE + K L+L F + + R D V C ++ + L + HE
Sbjct 73 IPALSALIALLNSDIPDIGETLAKELMLMFVQQFNRKDYVSCGNILQCLSILFLYDVIHE 132
> SPBC1734.04
Length=304
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 33/137 (24%), Positives = 58/137 (42%), Gaps = 16/137 (11%)
Query 50 LFRENLVRGRGLFCRALIRSQLASPGFT---GVYASLLCIVNSKLPDIGELVLKRLI-LQ 105
+FR+ +R G+ L+ F G + S N+ D+ E L++++ +
Sbjct 30 IFRQKTIRTVGIIALTLVLFLFFHRSFFSSFGEFPSFSSTANAPNSDVQEYDLRKVMNAK 89
Query 106 FRRAYRRNDKVVCLACVE--------FFAHLVNQRIAHELLALQLCELLLQQPTNDSVEV 157
F Y +KV+ A + FF H+ N HEL+ L L+ ++++EV
Sbjct 90 FTGDYSPKEKVLICAPLRNAAEHLNMFFGHMNNLTYPHELIDLA---FLISDTDDNTLEV 146
Query 158 CIGFLKAVGQVLEDVCR 174
L A+ Q ED +
Sbjct 147 LKQHLDAI-QNDEDESK 162
> SPAC24C9.11
Length=775
Score = 33.5 bits (75), Expect = 0.24, Method: Composition-based stats.
Identities = 38/161 (23%), Positives = 73/161 (45%), Gaps = 16/161 (9%)
Query 27 LRKSINGLVNKANVSNLTQLVQE---LFRENLVRG-----RGLFCRALIRSQLASPGFTG 78
LR+ + G +NK +++N++ +++E L+ EN L + ++ +
Sbjct 261 LRRKLQGSLNKLSIANISSIIKEIEVLYMENSRHSVTSTITNLLLQTVMGRESMLDQLAI 320
Query 79 VYASLLCIVNSKL-PDIGELVLKRLILQFRRAYRRNDKVVCLACVE------FFAHLVNQ 131
VYA+L + + D G +L+ L+ +F + Y+ +K + E FF L N
Sbjct 321 VYAALATALYRIVGNDFGAHLLQTLVERFLQLYKSKEKEPLSSHKETSNLIVFFVELYNF 380
Query 132 RIAHELLALQLCELLLQQPTNDSVEVCIGF-LKAVGQVLED 171
++ +L L L L+ T +VE+ + L GQ+ D
Sbjct 381 QLVSCVLVYDLIRLFLRSLTELNVEMLLKIVLNCGGQLRSD 421
> 7297773
Length=188
Score = 33.5 bits (75), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 8/61 (13%)
Query 85 CIVNSKLPDIGELVLKRLILQFRRAYRRNDKVVCLACVEFFAHLVNQRIAHELLALQLCE 144
C+ S + GELVL+ L F R +R+ +VCL C +F + + +L CE
Sbjct 15 CLATSSSVERGELVLEEL--PFARGPKRDSGIVCLGCYQFL------QFGEDGDSLDRCE 66
Query 145 L 145
L
Sbjct 67 L 67
> At1g57770
Length=578
Score = 31.6 bits (70), Expect = 0.83, Method: Composition-based stats.
Identities = 32/102 (31%), Positives = 49/102 (48%), Gaps = 11/102 (10%)
Query 1 PFKLAR-LQQQTSDSKSHAYQRQ--MWEALRKSINGL---VNKANVSNL-TQLVQELFRE 53
PF L L +++++ K+ QR MW A+ +++ GL K VS + T L + F
Sbjct 451 PFGLWEGLDRRSAEYKNLKSQRSEVMWRAVERAL-GLGFKREKCEVSLVGTPLTHQRF-- 507
Query 54 NLVRGRGLFCRALIRSQLASPGFTGVYASLLCIVNSKLPDIG 95
L R RG + A+ + PG + LLC +S P IG
Sbjct 508 -LRRNRGTYGPAIEAGKGTFPGHSTPIPQLLCCGDSTFPGIG 548
> CE27322
Length=496
Score = 31.6 bits (70), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 5/42 (11%)
Query 67 IRSQLASPGFTG-----VYASLLCIVNSKLPDIGELVLKRLI 103
IR QL G+ G ++ S LC + K P+IGE +K+L+
Sbjct 194 IREQLNEFGYPGDTCPVIFGSALCALEGKQPEIGEEAVKQLL 235
> CE23922
Length=410
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 49/113 (43%), Gaps = 2/113 (1%)
Query 33 GLVNKANVSNLTQLVQELFRENLVRGRGL-FCRALIRSQLASPGFTGVYASLLCIVNSKL 91
GLVN N ++Q LF R + L + + L +S + A L + S+
Sbjct 28 GLVNFGNTCYCNSVIQALFFCRPFREKVLNYKQTLKKSGASKDNLVTCLADLFHSIASQK 87
Query 92 PDIGELVLKRLILQFRRAYRRNDKVVCLACVEFFAHLVNQRIAHELLALQLCE 144
+G + KR I + ++ D + EFF +L+N I+ L+ ++ E
Sbjct 88 RRVGTIAPKRFITKLKKENELFDNYMQQDAHEFFNYLINT-ISETLIQEKIAE 139
> Hs12025673
Length=308
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 55/130 (42%), Gaps = 22/130 (16%)
Query 54 NLVRGRGLFCRAL-IRSQLASPGFTGVYASLL---CIVNSKLPDIGELVLKRLILQFRRA 109
+VR R L C+ L + +A F G ++ +V + LP + L+++ QF
Sbjct 173 GIVRLRTLLCKVLGVLFSVAGGLFVGKEGPMIHSGSVVGAGLPQFQSISLRKI--QFNFP 230
Query 110 YRRNDKVVCLACVEF----FAHLVNQRIAHELLA----------LQLCELLLQ-QPTNDS 154
Y R+D+ C +C F +LV R LL +C L Q P DS
Sbjct 231 YFRSDRSGCWSCCSFRGANRGYLVQSRGGFVLLEPRAHVESALLFHVCHLHPQLLPFWDS 290
Query 155 VEVCIGFLKA 164
V +GFL A
Sbjct 291 VWK-LGFLPA 299
> At3g60240_1
Length=1528
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 10/103 (9%)
Query 6 RLQQQTSDSKSHAYQRQMWEALRKSINGLVNKANVSNLTQLVQELFRENLVRGRGLFCRA 65
+ Q T + A QRQ+ KSI + N L + V+ + +N V G+ +
Sbjct 899 KYQVGTIADEEQAKQRQL-----KSILNKLTPQNFEKLFEQVKSVNIDNAVTLSGVISQI 953
Query 66 LIRSQLASPGFTGVYASLLCIVNSKLPDIGE----LVLKRLIL 104
++ L P F +YA ++ LPD E + KRL+L
Sbjct 954 FDKA-LMEPTFCEMYADFCFHLSGALPDFNENGEKITFKRLLL 995
> At5g63930
Length=1102
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 32 NGLVNKANVSNLTQLVQELFRENLVRGRGLFCRALIRSQLASPGFTG 78
+ L + NV+ + +L Q FR ++ R G C AL R QLA GFTG
Sbjct 475 SNLCKQVNVTAI-ELGQNRFRGSIPREVG-NCSALQRLQLADNGFTG 519
> At5g57870
Length=780
Score = 29.3 bits (64), Expect = 5.0, Method: Composition-based stats.
Identities = 40/183 (21%), Positives = 69/183 (37%), Gaps = 42/183 (22%)
Query 29 KSINGLVNKANVSNLTQLVQELFRENLVRG---RGLFCRALIRSQLASPGFTGVYASLLC 85
K++ G++NK L +L + +G+ ++ L P F +YA L
Sbjct 217 KTVKGILNKLTPEKYDLLKGQLIESGITSADILKGVITLIFDKAVL-EPTFCPMYAKLCS 275
Query 86 IVNSKLPDI-------GELVLKRLIL-----------QFRRAYR----------RND--- 114
+N +LP E+ KR++L Q R R RND
Sbjct 276 DINDQLPTFPPAEPGDKEITFKRVLLNICQEAFEGASQLREELRQMSAPDQEAERNDKEK 335
Query 115 --KVVCLACVEFFAHLVNQRIAHELLALQLCELLLQQ-----PTNDSVEVCIGFLKAVGQ 167
K+ L + L+ Q++ E + + + LL P ++VE F K +G+
Sbjct 336 LLKLKTLGNIRLIGELLKQKMVPEKIVHHIVQELLGADEKVCPAEENVEAICHFFKTIGK 395
Query 168 VLE 170
L+
Sbjct 396 QLD 398
> 7291420
Length=1303
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 3/41 (7%)
Query 19 YQRQM---WEALRKSINGLVNKANVSNLTQLVQELFRENLV 56
Y+R M +E LR+ I L KAN L + E+ R+NL+
Sbjct 582 YRRSMQVVYEKLRREIQELNEKANTQKLKEQSYEIKRKNLI 622
> At4g31540
Length=686
Score = 28.9 bits (63), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 49/113 (43%), Gaps = 13/113 (11%)
Query 71 LASPGFTGVYASLLCIVNSKLPDIGELVLKRL--ILQFRRAYRRNDKVVCLACVEFFAHL 128
+ASP G A CI S+LP V+ +L IL RA R DK C+ + +
Sbjct 210 MASPSSLGDQA---CIAPSQLP---VTVIHKLQAILGRLRANNRLDK-----CISIYVEV 258
Query 129 VNQRIAHELLALQLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAF 181
+ + L AL L L + + V+ G++ G LE + F+A F
Sbjct 259 RSLNVRASLQALDLDYLDISVSEFNDVQSIEGYIAQWGNHLEFAVKHLFEAEF 311
> CE08986
Length=170
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query 31 INGLVNKANVSNLTQLVQELFRENLVRGRGLFCRALIRSQLASPGFTGVY 80
++G+ + N ++LT +QE + E V F R + + A PG G Y
Sbjct 113 LHGIAEQRNDAHLTNYIQEKYLEEQVHSINEFARHIANIKRAGPGL-GEY 161
> CE06730
Length=3766
Score = 28.5 bits (62), Expect = 7.2, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 29/53 (54%), Gaps = 6/53 (11%)
Query 129 VNQRIAHELLALQLCELLLQQPTN----DSVEVCIGFLKAVGQVLEDVCRGGF 177
V + AH ++ + LC +L P N S+++ IG AV ++E++C G +
Sbjct 3200 VRDKAAHTVVTVHLCSSVLNAPQNWRVDKSIDMDIGRQTAV--LVENLCNGTY 3250
> 7304195
Length=336
Score = 28.1 bits (61), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 20/82 (24%), Positives = 37/82 (45%), Gaps = 9/82 (10%)
Query 101 RLILQFRRAYRRNDKV--VCLACVEFFAHLVNQRIAHELLA-------LQLCELLLQQPT 151
R+I Q ++ D+ VC+ EFF+ +R+ EL + + +LL Q +
Sbjct 212 RVITQVNEQFKNPDQTTFVCVCIAEFFSLYETERLVQELTKCGIDVHNIIVNQLLFLQNS 271
Query 152 NDSVEVCIGFLKAVGQVLEDVC 173
+DS +C K + L+ +
Sbjct 272 HDSCSMCASRFKIQEKYLDQIA 293
Lambda K H
0.327 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2842629034
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40