bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5329_orf1
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
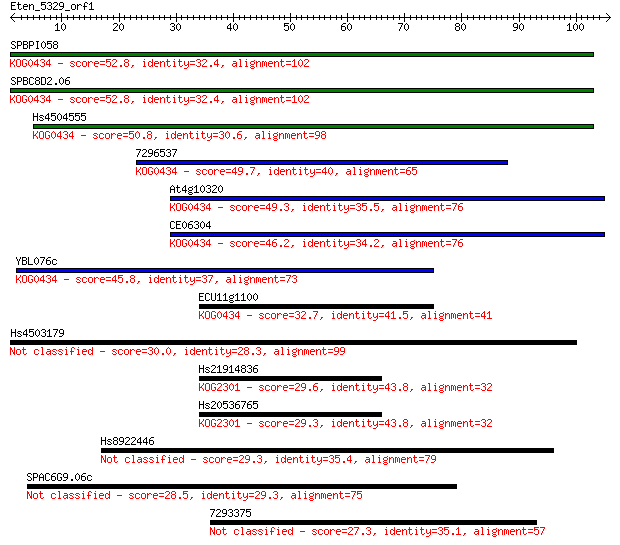
SPBPI058 52.8 2e-07
SPBC8D2.06 52.8 2e-07
Hs4504555 50.8 6e-07
7296537 49.7 1e-06
At4g10320 49.3 2e-06
CE06304 46.2 1e-05
YBL076c 45.8 2e-05
ECU11g1100 32.7 0.15
Hs4503179 30.0 0.95
Hs21914836 29.6 1.5
Hs20536765 29.3 1.6
Hs8922446 29.3 1.9
SPAC6G9.06c 28.5 3.4
7293375 27.3 6.8
> SPBPI058
Length=1064
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 33/104 (31%), Positives = 54/104 (51%), Gaps = 3/104 (2%)
Query 1 NGILLTPQEATLQRQLPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKL 60
+GI L + + R + + N L N D C++ +D D LQ + +ARE+ NRVQ+L
Sbjct 931 DGIELVEGDLQIIRSV-EVKNEFLKSNTDGICIVLLDIEIDAQLQAEGLAREVINRVQRL 989
Query 61 RKQLQLQQQDEV-LVFAAADDA-ALKDILIQEKEYIESCLRRPL 102
RK+ LQ D+V + + +D L+ + + LRRP+
Sbjct 990 RKKSNLQVTDDVRMTYKIKNDTIGLESAVDSNEALFSKVLRRPI 1033
> SPBC8D2.06
Length=1064
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 33/104 (31%), Positives = 54/104 (51%), Gaps = 3/104 (2%)
Query 1 NGILLTPQEATLQRQLPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKL 60
+GI L + + R + + N L N D C++ +D D LQ + +ARE+ NRVQ+L
Sbjct 931 DGIELVEGDLQIIRSV-EVKNEFLKSNTDGICIVLLDIEIDAQLQAEGLAREVINRVQRL 989
Query 61 RKQLQLQQQDEV-LVFAAADDA-ALKDILIQEKEYIESCLRRPL 102
RK+ LQ D+V + + +D L+ + + LRRP+
Sbjct 990 RKKSNLQVTDDVRMTYKIKNDTIGLESAVDSNEALFSKVLRRPI 1033
> Hs4504555
Length=1262
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 30/102 (29%), Positives = 53/102 (51%), Gaps = 4/102 (3%)
Query 5 LTPQEATLQRQLPQSTNPNLGF--NCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRK 62
L ++ L Q+T F + D ++ +D T D S+ + +ARE+ NR+QKLRK
Sbjct 940 LHDEDIRLMYTFDQATGGTAQFEAHSDAQALVLLDVTPDQSMVDEGMAREVINRIQKLRK 999
Query 63 QLQLQQQDEVLVF--AAADDAALKDILIQEKEYIESCLRRPL 102
+ L DE+ V+ A ++ L ++ E+I + ++ PL
Sbjct 1000 KCNLVPTDEITVYYKAKSEGTYLNSVIESHTEFIFTTIKAPL 1041
> 7296537
Length=1081
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 41/67 (61%), Gaps = 2/67 (2%)
Query 23 NLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVF--AAADD 80
N + D ++ +D T + L + +ARE+ NRVQKL+K+ QL D VL+F AAD+
Sbjct 972 NFEAHSDNEVLVLLDMTPNEELLEEGLAREVINRVQKLKKKAQLIPTDPVLIFHELAADN 1031
Query 81 AALKDIL 87
A +++L
Sbjct 1032 KAKQEVL 1038
> At4g10320
Length=1254
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 3/79 (3%)
Query 29 DKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAA---DDAALKD 85
D + ++ +D D SL AREI NR+QKLRK+ L+ D V V+ + D++ K
Sbjct 1006 DGDVLVILDLRADDSLVEAGFAREIVNRIQKLRKKSGLEPTDFVEVYFQSLDEDESVSKQ 1065
Query 86 ILIQEKEYIESCLRRPLLL 104
+L+ +++ I+ + LLL
Sbjct 1066 VLVSQEQNIKDSIGSTLLL 1084
> CE06304
Length=1141
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/78 (33%), Positives = 41/78 (52%), Gaps = 2/78 (2%)
Query 29 DKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQD--EVLVFAAADDAALKDI 86
D ++ +D TED SL + + RE+ NRVQ+LRKQ +L D V + + D+ L +
Sbjct 958 DAKTIVMIDTTEDESLIEEGLCREVTNRVQRLRKQAKLVSTDTAHVHIVVSPADSQLAKV 1017
Query 87 LIQEKEYIESCLRRPLLL 104
+ + I S P+ L
Sbjct 1018 VAAKLNDIVSATGTPIKL 1035
> YBL076c
Length=1072
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 40/74 (54%), Gaps = 1/74 (1%)
Query 2 GILLTPQEATLQRQLPQST-NPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKL 60
GI L + R LP+S D++ +I MD L+ + +ARE+ NR+QKL
Sbjct 939 GIELVKGDLNAIRGLPESAVQAGQETRTDQDVLIIMDTNIYSELKSEGLARELVNRIQKL 998
Query 61 RKQLQLQQQDEVLV 74
RK+ L+ D+VLV
Sbjct 999 RKKCGLEATDDVLV 1012
> ECU11g1100
Length=1017
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 34 IAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLV 74
I +D T D + + IARE + +QKLRK L+ D+V+V
Sbjct 931 IIIDNTLDEGMVKMKIAREFHSYIQKLRKSAGLRVGDDVVV 971
> Hs4503179
Length=1012
Score = 30.0 bits (66), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query 1 NGILLTPQEATLQRQLPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKL 60
N + P+++ + R + N N+ C N I+ DF +P Q +AR +A+R+
Sbjct 555 NEVYCNPKQSVIDRSVNGLINGNV-VPC--NGEISGDFLNNPFKQENVLARMVASRITNY 611
Query 61 -RKQLQLQQQDEVLVFAAADDAALKDILIQEKEYIESCLR 99
++ D L F A A +K+ L QE E +E R
Sbjct 612 PTAWVEGSSPDSDLEFVANTKARVKE-LQQEAERLEKAFR 650
> Hs21914836
Length=1999
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 2/34 (5%)
Query 34 IAMDFTEDPSLQRKA--IAREIANRVQKLRKQLQ 65
++MDF EDPS +++A IA + N V++L + Q
Sbjct 688 VSMDFLEDPSQRQRAMSIASILTNTVEELEESRQ 721
> Hs20536765
Length=1649
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 2/34 (5%)
Query 34 IAMDFTEDPSLQRKA--IAREIANRVQKLRKQLQ 65
++MDF EDPS +++A IA + N V++L + Q
Sbjct 339 VSMDFLEDPSQRQRAMSIASILTNTVEELEESRQ 372
> Hs8922446
Length=213
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 42/92 (45%), Gaps = 19/92 (20%)
Query 17 PQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQ----------- 65
P T NL CD N + ED QRK I ++ +V+KLRK+L+
Sbjct 122 PLQTPVNLSVFCDHNYTV-----EDTMHQRKRI-HQLEQQVEKLRKKLKTAQQRCRRQER 175
Query 66 -LQQQDEVLVF-AAADDAALKDILIQEKEYIE 95
L++ EV+ F DD + + +I +Y E
Sbjct 176 QLEKLKEVVHFQKEKDDVSERGYVILPNDYFE 207
> SPAC6G9.06c
Length=1208
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 44/78 (56%), Gaps = 5/78 (6%)
Query 4 LLTPQE---ATLQRQLPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKL 60
LLT +E ATL+RQ+ + N + F ++N + ED ++ +A E A+R+Q L
Sbjct 307 LLTEKEDEIATLKRQIEEKENSSSAFENEENSSY-VHLQEDYAI-LQAKCDEFADRIQVL 364
Query 61 RKQLQLQQQDEVLVFAAA 78
L+ +++++++ + A
Sbjct 365 TADLEKEKENQIMHESEA 382
> 7293375
Length=1187
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query 36 MDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAADDAALKDILIQEKE 92
MD L+R +++R++KLR+ Q QQQ EV + A DA K L ++E
Sbjct 166 MDKDGPTDLERPGDWDSLSDRMRKLRRS-QQQQQKEVALLADRKDALTKRPLDFDRE 221
Lambda K H
0.320 0.134 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1170944580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40