bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
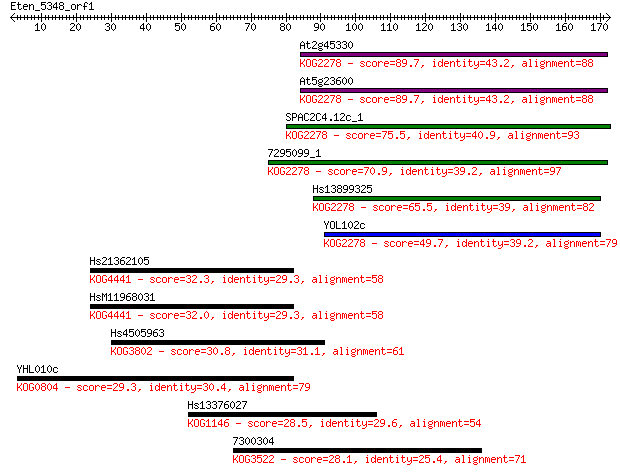
Query= Eten_5348_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
At2g45330 89.7 2e-18
At5g23600 89.7 3e-18
SPAC2C4.12c_1 75.5 4e-14
7295099_1 70.9 1e-12
Hs13899325 65.5 4e-11
YOL102c 49.7 3e-06
Hs21362105 32.3 0.46
HsM11968031 32.0 0.62
Hs4505963 30.8 1.4
YHL010c 29.3 4.3
Hs13376027 28.5 6.3
7300304 28.1 9.0
> At2g45330
Length=257
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 59/88 (67%), Gaps = 0/88 (0%)
Query 84 SGKWLLRATQGHTIPYIRSDLFMRRVENAAELPICAHGTYLRNWMAIRHLGLHRMHRNHI 143
+G+ L+RA QGH+I + S+ ++ + + E P+C HGTY +N +I GL RM+R H+
Sbjct 124 NGELLIRANQGHSITTVESEKLLKPILSPEEAPVCVHGTYRKNLESILASGLKRMNRMHV 183
Query 144 HFATGLPGEATVLSGMRRNSEVAVYLHV 171
HF+ GLP + V+SGMRRN V ++L +
Sbjct 184 HFSCGLPTDGEVISGMRRNVNVIIFLDI 211
> At5g23600
Length=212
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 59/88 (67%), Gaps = 0/88 (0%)
Query 84 SGKWLLRATQGHTIPYIRSDLFMRRVENAAELPICAHGTYLRNWMAIRHLGLHRMHRNHI 143
G+ L+RA QGH+I + S+ ++ + + E P+C HGTY +N +I GL RM+R H+
Sbjct 79 DGELLIRANQGHSITTVESEKLLKPILSPEEAPVCVHGTYRKNLESILASGLKRMNRMHV 138
Query 144 HFATGLPGEATVLSGMRRNSEVAVYLHV 171
HF+ GLP + V+SG+RRN V ++LH+
Sbjct 139 HFSCGLPTDGEVISGVRRNVNVIIFLHI 166
> SPAC2C4.12c_1
Length=225
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 38/93 (40%), Positives = 57/93 (61%), Gaps = 2/93 (2%)
Query 80 VERLSGKWLLRATQGHTIPYIRSDLFMRRVENAAELPICAHGTYLRNWMAIRHLGLHRMH 139
+E + G +RA QGH+I ++ + M R++NA+ +P HGT W I GL RM
Sbjct 95 MEEVEGVLYIRANQGHSIKAVQ--VPMARIDNASSIPKVVHGTKKELWPVISKQGLSRMK 152
Query 140 RNHIHFATGLPGEATVLSGMRRNSEVAVYLHVA 172
RNHIH ATGL G+ V+SG+R++ + +Y+ A
Sbjct 153 RNHIHCATGLYGDPGVISGIRKSCTLYIYIDSA 185
> 7295099_1
Length=245
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 38/105 (36%), Positives = 62/105 (59%), Gaps = 9/105 (8%)
Query 75 AAAKKVERLSGKW-------LLRATQGHTIPYIRSDLF-MRRVENAAELPICAHGTYLRN 126
AAA +R + +W +RA QGH++ + + + + + +++P+ HGTY R+
Sbjct 59 AAADAKQRYTLRWNPELGVHEIRANQGHSLAVLEGEAGGLENITHVSQVPLAVHGTYYRH 118
Query 127 WMAIRHLGLHRMHRNHIHFATGLPGEATVLSGMRRNSEVAVYLHV 171
W AIR GL RM+RNH+HFA +T LSG R + ++ +YL+V
Sbjct 119 WGAIRSQGLSRMNRNHVHFACSDETNST-LSGFRSDCQILIYLNV 162
> Hs13899325
Length=204
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 49/82 (59%), Gaps = 1/82 (1%)
Query 88 LLRATQGHTIPYIRSDLFMRRVENAAELPICAHGTYLRNWMAIRHLGLHRMHRNHIHFAT 147
L+RA QGH++ + +L A P+ HGT+ ++W +I GL R HIH A
Sbjct 50 LIRANQGHSLQVPKLELMPLETPQALP-PMLVHGTFWKHWPSILLKGLSCQGRTHIHLAP 108
Query 148 GLPGEATVLSGMRRNSEVAVYL 169
GLPG+ ++SGMR + E+AV++
Sbjct 109 GLPGDPGIISGMRSHCEIAVFI 130
> YOL102c
Length=230
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 49/82 (59%), Gaps = 3/82 (3%)
Query 91 ATQGHTIPYIR-SDLFMRRVENAAELPI-CAHGTYLRNWMAIRHLG-LHRMHRNHIHFAT 147
ATQGH+I I+ SD + + A++LP HGT L++ + I G + M RNH+H +
Sbjct 86 ATQGHSIKSIQPSDEVLVPITEASQLPQELIHGTNLQSVIKIIESGAISPMSRNHVHLSP 145
Query 148 GLPGEATVLSGMRRNSEVAVYL 169
G+ V+SGMR +S V +++
Sbjct 146 GMLHAKGVISGMRSSSNVYIFI 167
> Hs21362105
Length=589
Score = 32.3 bits (72), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 24/58 (41%), Gaps = 0/58 (0%)
Query 24 ERSRCSSGCSSSGGLTTPPAAAAGAAGESLCCKSGSTVVCGSVACADHGDKAAAKKVE 81
E RC + + G+ T P A AG +L G T +C + DH K K +
Sbjct 268 EALRCKTRILQNDGVVTSPCARPRKAGHTLLILGGQTFMCDKIYQVDHKAKEIIPKAD 325
> HsM11968031
Length=438
Score = 32.0 bits (71), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 24/58 (41%), Gaps = 0/58 (0%)
Query 24 ERSRCSSGCSSSGGLTTPPAAAAGAAGESLCCKSGSTVVCGSVACADHGDKAAAKKVE 81
E RC + + G+ T P A AG +L G T +C + DH K K +
Sbjct 117 EALRCKTRILQNDGVVTSPCARPRKAGHTLLILGGQTFMCDKIYQVDHKAKEIIPKAD 174
> Hs4505963
Length=361
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query 30 SGCSSSGGLTTPPAAAAGAAGESLCCKSGSTVVCGSVACADHGDK--AAAKKVERLSGKW 87
SG GGLT PPAAA+ + + + GS C DH D+ + ++E+ + ++
Sbjct 142 SGMLEHGGLTPPPAAASAQSLHPVLREPPDHGELGSHHCQDHSDEETPTSDELEQFAKQF 201
Query 88 LLR 90
R
Sbjct 202 KQR 204
> YHL010c
Length=585
Score = 29.3 bits (64), Expect = 4.3, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 35/91 (38%), Gaps = 13/91 (14%)
Query 3 EQQADSTQDLAALGC-----CDCAHA-ERSRCSSGCSSSGGLTTPPAAAAGAAGESLCCK 56
E+ T L + C C C + + SRC C S + + A + C
Sbjct 245 ERMDSETTGLVTIPCQHTFHCQCLNKWKNSRCPV-CRHSSLRLSRESLLKQAGDSAHCAT 303
Query 57 SGST------VVCGSVACADHGDKAAAKKVE 81
GST ++CG+V C + K A K E
Sbjct 304 CGSTDNLWICLICGNVGCGRYNSKHAIKHYE 334
> Hs13376027
Length=324
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 25/58 (43%), Gaps = 4/58 (6%)
Query 52 SLCCKSGSTVVCGSVACADHGDKAAAKKVERLSGKWLLRATQGH----TIPYIRSDLF 105
SLC SG + +C+D D ++K+E L ++A IR D+F
Sbjct 265 SLCSTSGVQTSLPTESCSDESDSELSQKLEDLDNSLEVKAKPASGLDGNFNSIRMDMF 322
> 7300304
Length=1101
Score = 28.1 bits (61), Expect = 9.0, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 32/71 (45%), Gaps = 1/71 (1%)
Query 65 SVACADHGDKAAAKKVERLSGKWLLRATQGHTIPYIRSDLFMRRVENAAELPICAHGTYL 124
S+ C+D D + + +S KW L +G T Y+ +++ R ++ E YL
Sbjct 123 SITCSDSEDDSERRAQFEISSKWNLGGYEGDTRTYVVQEIY-RNEQSYVESLQTVVVKYL 181
Query 125 RNWMAIRHLGL 135
+ A H G+
Sbjct 182 KVLKAPEHAGM 192
Lambda K H
0.320 0.131 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40