bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5401_orf1
Length=135
Score E
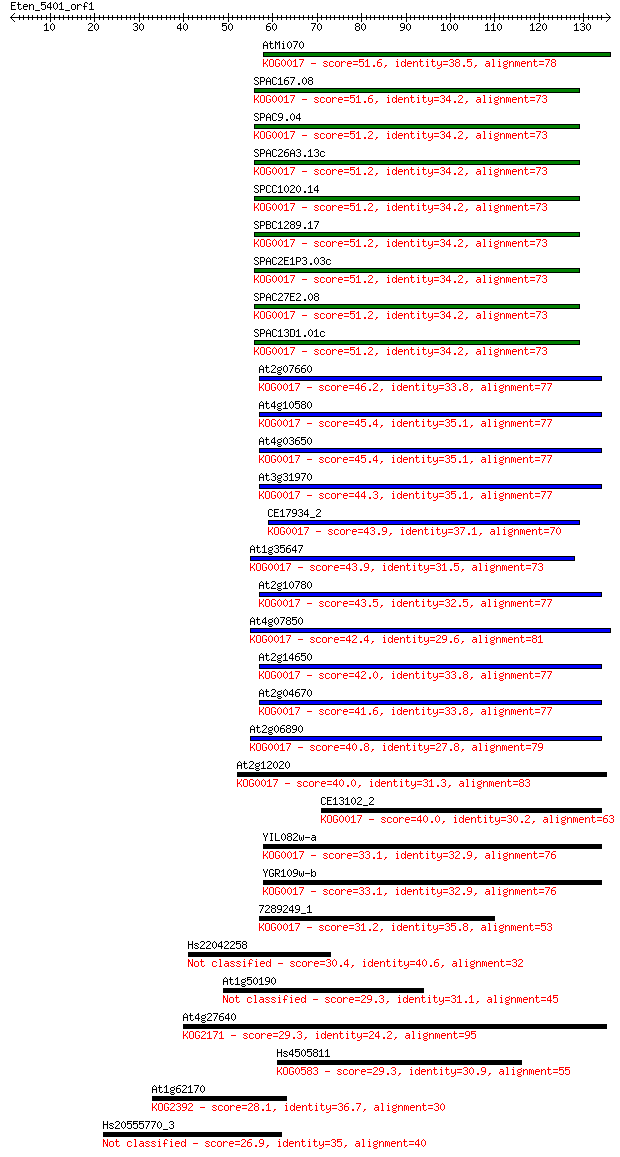
Sequences producing significant alignments: (Bits) Value
AtMi070 51.6 4e-07
SPAC167.08 51.6 5e-07
SPAC9.04 51.2 5e-07
SPAC26A3.13c 51.2 5e-07
SPCC1020.14 51.2 5e-07
SPBC1289.17 51.2 5e-07
SPAC2E1P3.03c 51.2 5e-07
SPAC27E2.08 51.2 5e-07
SPAC13D1.01c 51.2 5e-07
At2g07660 46.2 2e-05
At4g10580 45.4 3e-05
At4g03650 45.4 3e-05
At3g31970 44.3 7e-05
CE17934_2 43.9 8e-05
At1g35647 43.9 9e-05
At2g10780 43.5 1e-04
At4g07850 42.4 2e-04
At2g14650 42.0 3e-04
At2g04670 41.6 4e-04
At2g06890 40.8 7e-04
At2g12020 40.0 0.001
CE13102_2 40.0 0.001
YIL082w-a 33.1 0.14
YGR109w-b 33.1 0.17
7289249_1 31.2 0.51
Hs22042258 30.4 0.94
At1g50190 29.3 2.3
At4g27640 29.3 2.4
Hs4505811 29.3 2.4
At1g62170 28.1 4.9
Hs20555770_3 26.9 9.5
> AtMi070
Length=158
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 43/78 (55%), Gaps = 3/78 (3%)
Query 58 KIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELHT 117
K+EA+ WPE +N T++R FLG Y F+ NY + RPL + +KN S KWTE+
Sbjct 50 KLEAMVGWPEP-KNTTELRGFLGLTGYYRRFV-KNYGKIVRPLTELLKKN-SLKWTEMAA 106
Query 118 QAVRQLKQRLIDFTTLQV 135
A + LK + L +
Sbjct 107 LAFKALKGAVTTLPVLAL 124
> SPAC167.08
Length=1214
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 505 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 562
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 563 QTQAIENIKQCLV 575
> SPAC9.04
Length=1333
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 624 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 681
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 682 QTQAIENIKQCLV 694
> SPAC26A3.13c
Length=1333
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 624 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 681
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 682 QTQAIENIKQCLV 694
> SPCC1020.14
Length=1333
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 624 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 681
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 682 QTQAIENIKQCLV 694
> SPBC1289.17
Length=1333
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 624 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 681
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 682 QTQAIENIKQCLV 694
> SPAC2E1P3.03c
Length=1333
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 624 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 681
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 682 QTQAIENIKQCLV 694
> SPAC27E2.08
Length=1333
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 624 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 681
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 682 QTQAIENIKQCLV 694
> SPAC13D1.01c
Length=1333
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 56 KDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
++ I+ + W + +N ++RQFLG++NY F+ P + + PL + +K+V +KWT
Sbjct 624 QENIDKVLQWKQP-KNRKELRQFLGSVNYLRKFI-PKTSQLTHPLNNLLKKDVRWKWTPT 681
Query 116 HTQAVRQLKQRLI 128
TQA+ +KQ L+
Sbjct 682 QTQAIENIKQCLV 694
> At2g07660
Length=949
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 43/77 (55%), Gaps = 2/77 (2%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELH 116
+KI +I WP N T++R FLG Y F+ ++A +A+PL T K+ +F W++
Sbjct 367 EKIRSIKEWPRP-RNATEIRSFLGLAGYYRRFVM-SFASMAQPLTRLTGKDTAFNWSDEC 424
Query 117 TQAVRQLKQRLIDFTTL 133
++ +LK LI+ L
Sbjct 425 EKSFLELKAMLINAPVL 441
> At4g10580
Length=1240
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 40/77 (51%), Gaps = 2/77 (2%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELH 116
+KIEAI WP N T++R FLG Y F+ +A +A+P+ T K+V F W++
Sbjct 704 EKIEAIRDWPRPT-NATEIRSFLGWAGYYRRFVK-GFASMAQPMTKLTGKDVPFVWSQEC 761
Query 117 TQAVRQLKQRLIDFTTL 133
+ LK+ L L
Sbjct 762 EEGFVSLKEMLTSTPVL 778
> At4g03650
Length=839
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 39/77 (50%), Gaps = 2/77 (2%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELH 116
+KIEAI WP N T++R FLG Y F+ +A +A+P+ T K+V F W+
Sbjct 629 EKIEAIRDWPRPT-NATEIRSFLGLAGYYRRFI-KGFASMAQPMTKLTGKDVPFVWSPEC 686
Query 117 TQAVRQLKQRLIDFTTL 133
+ LK+ L L
Sbjct 687 EEGFVSLKEMLTSTPVL 703
> At3g31970
Length=1329
Score = 44.3 bits (103), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 39/77 (50%), Gaps = 2/77 (2%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELH 116
+KIEAI WP N T++R FLG Y F+ +A +A+P+ T K+V F W+
Sbjct 664 EKIEAIRDWPRPT-NATEIRSFLGLAGYYKRFV-KVFASMAQPMTKLTGKDVPFVWSPEC 721
Query 117 TQAVRQLKQRLIDFTTL 133
+ LK+ L L
Sbjct 722 DEGFMSLKEMLTSTPVL 738
> CE17934_2
Length=696
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 2/70 (2%)
Query 59 IEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELHTQ 118
++AI + P N VR+F+G + F+ PN++++A PL TRK F W +
Sbjct 156 VDAIVNLP-TPRNVGDVRRFIGMSGFFRKFL-PNFSEIAEPLTRFTRKGHKFVWKAEQQK 213
Query 119 AVRQLKQRLI 128
AV LKQ LI
Sbjct 214 AVDTLKQALI 223
> At1g35647
Length=1495
Score = 43.9 bits (102), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 42/73 (57%), Gaps = 2/73 (2%)
Query 55 SKDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTE 114
++K++AI WP ++ +VR F G + F+ +++ +A PL + +KNV FKW +
Sbjct 729 DEEKVKAIREWPSP-KSVGEVRSFHGLAGFYRRFVK-DFSTLAAPLTEVIKKNVGFKWEQ 786
Query 115 LHTQAVRQLKQRL 127
A + LK++L
Sbjct 787 AQEDAFQALKEKL 799
> At2g10780
Length=1611
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 42/77 (54%), Gaps = 2/77 (2%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELH 116
+KI +I WP N T++R FLG Y F+ ++A +A+PL T K+ +F W++
Sbjct 872 EKIRSIKEWPRP-RNATEIRSFLGLAGYYRRFVM-SFASMAQPLTRLTGKDTAFNWSDEC 929
Query 117 TQAVRQLKQRLIDFTTL 133
++ +LK L + L
Sbjct 930 EKSFLELKAMLTNAPVL 946
> At4g07850
Length=1138
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Query 55 SKDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTE 114
++K++AI WP ++ +VR F G + F+ +++ +A PL + +KNV FKW +
Sbjct 540 DEEKVKAIREWPSP-KSVGKVRSFHGLAGFYRRFVR-DFSTLAAPLTEVIKKNVGFKWEQ 597
Query 115 LHTQAVRQLKQRLIDFTTLQV 135
A + LK++L L +
Sbjct 598 APEDAFQALKEKLTHAPVLSL 618
> At2g14650
Length=1328
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELH 116
+KIEAI W N T++R FLG Y F+ +A +A+P+ T K+V F W+
Sbjct 679 EKIEAIRDW-HTPTNATEIRSFLGLAGYYRRFV-KGFASMAQPMTKLTGKDVPFVWSPEC 736
Query 117 TQAVRQLKQRLIDFTTL 133
+ LK+ L L
Sbjct 737 EEGFVSLKEMLTSTPVL 753
> At2g04670
Length=1411
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELH 116
+KIEAI WP N T++R FL Y F+ +A +A+P+ T K+V F W+
Sbjct 730 EKIEAIRDWPRPT-NATEIRSFLRLTGYYRRFVK-GFASMAQPMTKLTGKDVPFVWSPEC 787
Query 117 TQAVRQLKQRLIDFTTL 133
+ LK+ L L
Sbjct 788 EEGFVSLKEMLTSTPVL 804
> At2g06890
Length=1215
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query 55 SKDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTE 114
++K++AI WP + +VR F G + F +++ + PL + +K+V FKW +
Sbjct 630 DEEKVKAIRDWPSP-KTVGEVRSFHGLAGFYRRFFK-DFSTIVAPLTEVMKKDVGFKWEK 687
Query 115 LHTQAVRQLKQRLIDFTTL 133
+A + LK +L + L
Sbjct 688 AQEEAFQSLKDKLTNAPVL 706
> At2g12020
Length=976
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 43/83 (51%), Gaps = 2/83 (2%)
Query 52 LRGSKDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFK 111
+ + KIE + + L+N VR FLG + F+ +++ +ARPL K V F+
Sbjct 153 IEADRAKIEVMTSL-QALDNLKAVRSFLGHAGFYRRFIK-DFSKIARPLTSLLCKEVKFE 210
Query 112 WTELHTQAVRQLKQRLIDFTTLQ 134
+T+ A +Q+KQ LI +Q
Sbjct 211 FTQECHDAFQQIKQALISAPIVQ 233
> CE13102_2
Length=813
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 36/63 (57%), Gaps = 1/63 (1%)
Query 71 NETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTELHTQAVRQLKQRLIDF 130
++ Q+ FLG +++ S F+ P + + PL +++V + WT++H +A LK + D
Sbjct 268 DQKQLASFLGAVSFYSRFV-PKMSKLRGPLDSLMKRDVKWNWTDIHQEAFNTLKNAVADS 326
Query 131 TTL 133
T L
Sbjct 327 TML 329
> YIL082w-a
Length=1498
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 40/80 (50%), Gaps = 12/80 (15%)
Query 58 KIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARP----LVDHTRKNVSFKWT 113
K AI +P + Q ++FLG INY F+ PN + +A+P + D + +WT
Sbjct 834 KCAAIRDFP-TPKTVKQAQRFLGMINYYRRFI-PNCSKIAQPIQLFICDKS------QWT 885
Query 114 ELHTQAVRQLKQRLIDFTTL 133
E +A+ +LK L + L
Sbjct 886 EKQDKAIEKLKAALCNSPVL 905
> YGR109w-b
Length=1547
Score = 33.1 bits (74), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 40/80 (50%), Gaps = 12/80 (15%)
Query 58 KIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARP----LVDHTRKNVSFKWT 113
K AI +P + Q ++FLG INY F+ PN + +A+P + D + +WT
Sbjct 808 KCAAIRDFP-TPKTVKQAQRFLGMINYYRRFI-PNCSKIAQPIQLFICDKS------QWT 859
Query 114 ELHTQAVRQLKQRLIDFTTL 133
E +A+ +LK L + L
Sbjct 860 EKQDKAIDKLKDALCNSPVL 879
> 7289249_1
Length=276
Score = 31.2 bits (69), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Query 57 DKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVS 109
+KI+ I + V +N+ + R FLG I Y F+ P+ A+ PL RK +S
Sbjct 19 EKIQIINSF-RVPKNKEETRSFLGLITYVGKFI-PDLAENTEPLRQLLRKALS 69
> Hs22042258
Length=546
Score = 30.4 bits (67), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 41 KQLRIWVIPFLLRGSKDKIEAIGHWPEVLENE 72
++LR W + ++RGS D+ E IG P NE
Sbjct 142 RKLRDWSLEQMVRGSSDQPEDIGQSPSGTTNE 173
> At1g50190
Length=637
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 14/57 (24%), Positives = 28/57 (49%), Gaps = 12/57 (21%)
Query 49 PFLLRGSKDKIEAIGHWPEVLENETQVRQFLGTINYCS------------MFMGPNY 93
P L ++K + + +W E+ E+E +++ T +YCS ++M PN+
Sbjct 467 PLTLCCGEEKTKDLQYWCEICESEIDAQKWFYTCSYCSVTLHVTCLLGDKVYMKPNH 523
> At4g27640
Length=651
Score = 29.3 bits (64), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/101 (22%), Positives = 46/101 (45%), Gaps = 18/101 (17%)
Query 40 GKQLRIWVIPFLL------RGSKDKIEAIGHWPEVLENETQVRQFLGTINYCSMFMGPNY 93
+ L + +I FL+ R ++D+I+ + P+V+ + + PN
Sbjct 2 AQSLELLLIQFLMPDNDARRQAEDQIKRLAKDPQVVP---------ALVQHLRTAKTPNV 52
Query 94 ADVARPLVDHTRKNVSFKWTELHTQAVRQLKQRLIDFTTLQ 134
+A L+ RK ++ W +L Q + +KQ LI+ T++
Sbjct 53 RQLAAVLL---RKRITGHWAKLSPQLKQHVKQSLIESITVE 90
> Hs4505811
Length=313
Score = 29.3 bits (64), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 61 AIGHWPEVLENETQVRQFLGTINYCSMFMGPNYADVARPLVDHTRKNVSFKWTEL 115
A G E LE++ QV LG+ + S++ G +D + H K+ W EL
Sbjct 26 APGKEKEPLESQYQVGPLLGSGGFGSVYSGIRVSDNLPVAIKHVEKDRISDWGEL 80
> At1g62170
Length=433
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 33 SANGSLQGKQLRIWVIPFLLRGSKDKIEAI 62
+++G QG++LR +++ FL S D++ AI
Sbjct 108 ASSGGEQGEELRSFILSFLKSSSTDELNAI 137
> Hs20555770_3
Length=591
Score = 26.9 bits (58), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 22 AFFLEISSTQNSANGSLQGKQLRIWVIPFLLRGSKDKIEA 61
A+ +I S N N S+QGK + + + G K K+EA
Sbjct 369 AYLSDIFSIFNDLNASMQGKNATYFSMADKVEGQKQKLEA 408
Lambda K H
0.323 0.136 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40