bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5444_orf2
Length=149
Score E
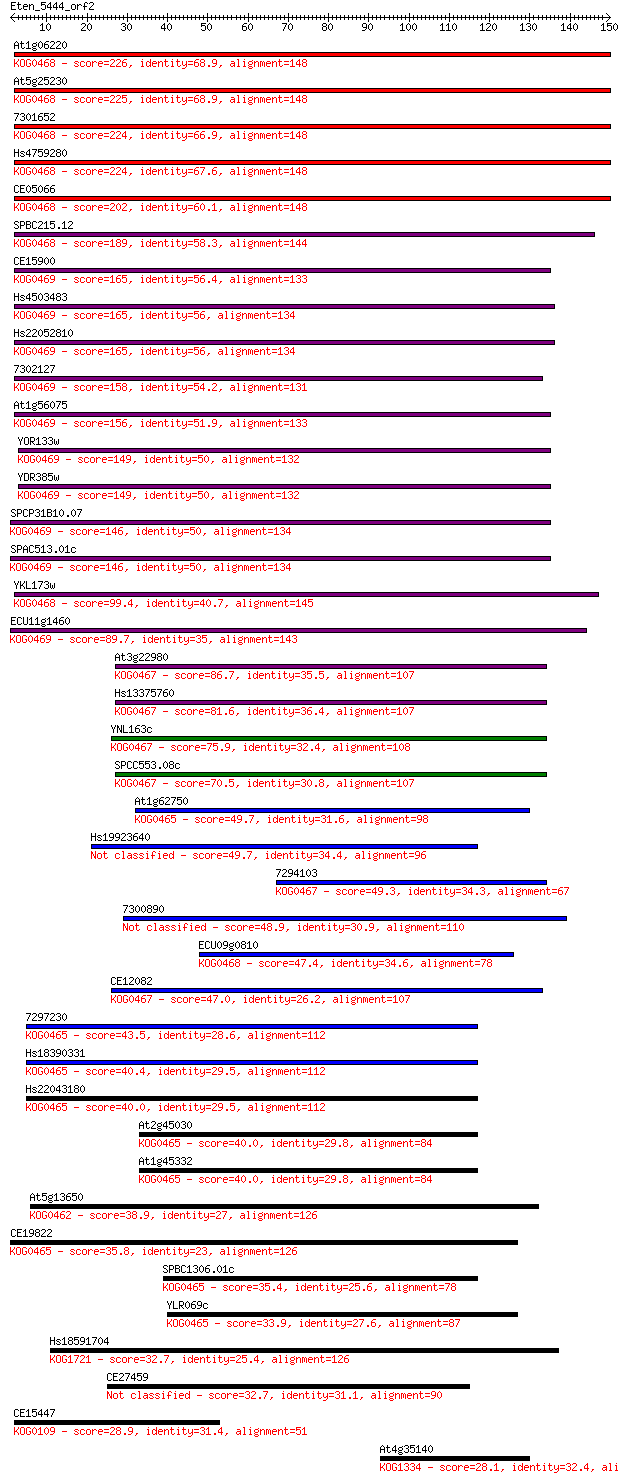
Sequences producing significant alignments: (Bits) Value
At1g06220 226 1e-59
At5g25230 225 2e-59
7301652 224 4e-59
Hs4759280 224 4e-59
CE05066 202 2e-52
SPBC215.12 189 1e-48
CE15900 165 2e-41
Hs4503483 165 4e-41
Hs22052810 165 4e-41
7302127 158 3e-39
At1g56075 156 1e-38
YOR133w 149 2e-36
YDR385w 149 2e-36
SPCP31B10.07 146 1e-35
SPAC513.01c 146 1e-35
YKL173w 99.4 2e-21
ECU11g1460 89.7 2e-18
At3g22980 86.7 2e-17
Hs13375760 81.6 5e-16
YNL163c 75.9 3e-14
SPCC553.08c 70.5 1e-12
At1g62750 49.7 2e-06
Hs19923640 49.7 2e-06
7294103 49.3 3e-06
7300890 48.9 4e-06
ECU09g0810 47.4 1e-05
CE12082 47.0 1e-05
7297230 43.5 2e-04
Hs18390331 40.4 0.001
Hs22043180 40.0 0.001
At2g45030 40.0 0.002
At1g45332 40.0 0.002
At5g13650 38.9 0.004
CE19822 35.8 0.028
SPBC1306.01c 35.4 0.045
YLR069c 33.9 0.13
Hs18591704 32.7 0.26
CE27459 32.7 0.28
CE15447 28.9 3.9
At4g35140 28.1 5.8
> At1g06220
Length=987
Score = 226 bits (577), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 102/148 (68%), Positives = 124/148 (83%), Gaps = 0/148 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
+E ++ V+FK+VDA IA + +HRG GQ+IP+ARRVAYSA L+ATPRL+EP+ + EIQ P
Sbjct 795 DEPIRNVKFKIVDARIAPEPLHRGSGQMIPTARRVAYSAFLMATPRLMEPVYYVEIQTPI 854
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
DCV+AIYTVL+RRRG+V D+P+PGTP YIV A+LP IESFGFETDLR HT GQA CL+V
Sbjct 855 DCVTAIYTVLSRRRGHVTSDVPQPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSV 914
Query 122 FDNWAIVPGDPLDKQIVFKPLEPAPAPH 149
FD+WAIVPGDPLDK I +PLEPAP H
Sbjct 915 FDHWAIVPGDPLDKAIQLRPLEPAPIQH 942
> At5g25230
Length=973
Score = 225 bits (574), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 102/148 (68%), Positives = 123/148 (83%), Gaps = 0/148 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
+E ++ V+FK+VDA IA + +HRG GQ+IP+ARRVAYSA L+ATPRL+EP+ + EIQ P
Sbjct 781 DEPIRNVKFKIVDARIAPEPLHRGSGQMIPTARRVAYSAFLMATPRLMEPVYYVEIQTPI 840
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
DCV+AIYTVL+RRRG V D+P+PGTP YIV A+LP IESFGFETDLR HT GQA CL+V
Sbjct 841 DCVTAIYTVLSRRRGYVTSDVPQPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSV 900
Query 122 FDNWAIVPGDPLDKQIVFKPLEPAPAPH 149
FD+WAIVPGDPLDK I +PLEPAP H
Sbjct 901 FDHWAIVPGDPLDKAIQLRPLEPAPIQH 928
> 7301652
Length=975
Score = 224 bits (572), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 99/148 (66%), Positives = 124/148 (83%), Gaps = 0/148 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
EE ++ V+FK++D IA +A+HRGGGQIIP+ARRVAYSA L+ATPRL+EP +F E+Q PA
Sbjct 784 EEPIRNVKFKILDGVIANEALHRGGGQIIPTARRVAYSAFLMATPRLMEPYLFVEVQAPA 843
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
DCVSA+YTVLARRRG+V +D P G+P+Y + A++PAI+SFGFETDLRTHT GQA CL+V
Sbjct 844 DCVSAVYTVLARRRGHVTQDAPVSGSPIYTIKAFIPAIDSFGFETDLRTHTQGQAFCLSV 903
Query 122 FDNWAIVPGDPLDKQIVFKPLEPAPAPH 149
F +W IVPGDPLDK I+ +PLEP A H
Sbjct 904 FHHWQIVPGDPLDKSIIIRPLEPQQASH 931
> Hs4759280
Length=972
Score = 224 bits (572), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 100/148 (67%), Positives = 125/148 (84%), Gaps = 0/148 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
+E ++ V+FK++DA +A++ +HRGGGQIIP+ARRV YSA L+ATPRL+EP F E+Q PA
Sbjct 781 DELIRNVKFKILDAVVAQEPLHRGGGQIIPTARRVVYSAFLMATPRLMEPYYFVEVQAPA 840
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
DCVSA+YTVLARRRG+V +D P PG+PLY + A++PAI+SFGFETDLRTHT GQA L+V
Sbjct 841 DCVSAVYTVLARRRGHVTQDAPIPGSPLYTIKAFIPAIDSFGFETDLRTHTQGQAFSLSV 900
Query 122 FDNWAIVPGDPLDKQIVFKPLEPAPAPH 149
F +W IVPGDPLDK IV +PLEP PAPH
Sbjct 901 FHHWQIVPGDPLDKSIVIRPLEPQPAPH 928
> CE05066
Length=974
Score = 202 bits (514), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 89/148 (60%), Positives = 116/148 (78%), Gaps = 0/148 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
EE ++ V+FKL+DA IA + ++RGGGQ+IP+ARR AYSA L+ATPRL+EP E+ PA
Sbjct 785 EEPIRQVKFKLLDAAIATEPLYRGGGQMIPTARRCAYSAFLMATPRLMEPYYTVEVVAPA 844
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
DCV+A+YTVLA+RRG+V D P PG+P+Y + AY+P ++SFGFETDLR HT GQA C++
Sbjct 845 DCVAAVYTVLAKRRGHVTTDAPMPGSPMYTISAYIPVMDSFGFETDLRIHTQGQAFCMSA 904
Query 122 FDNWAIVPGDPLDKQIVFKPLEPAPAPH 149
F +W +VPGDPLDK IV K L+ P PH
Sbjct 905 FHHWQLVPGDPLDKSIVIKTLDVQPTPH 932
> SPBC215.12
Length=983
Score = 189 bits (481), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 84/144 (58%), Positives = 111/144 (77%), Gaps = 0/144 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
+E ++ V F+L+D +A + ++RGGGQIIP+ARRV YS+ L A+PRL+EP+ E+ PA
Sbjct 794 DETIRNVNFRLMDVVLAPEQIYRGGGQIIPTARRVCYSSFLTASPRLMEPVYMVEVHAPA 853
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
D + IY +L RRRG+V +D+P+PG+PLY+V A +P I+S GFETDLR HT GQAMC V
Sbjct 854 DSLPIIYDLLTRRRGHVLQDIPRPGSPLYLVRALIPVIDSCGFETDLRVHTQGQAMCQMV 913
Query 122 FDNWAIVPGDPLDKQIVFKPLEPA 145
FD+W +VPGDPLDK I KPLEPA
Sbjct 914 FDHWQVVPGDPLDKSIKPKPLEPA 937
> CE15900
Length=852
Score = 165 bits (418), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 75/133 (56%), Positives = 97/133 (72%), Gaps = 0/133 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
+EN++GVRF + D + DA+HRGGGQIIP+ARRV Y+++L A PRLLEP+ EIQCP
Sbjct 688 DENMRGVRFNVHDVTLHADAIHRGGGQIIPTARRVFYASVLTAEPRLLEPVYLVEIQCPE 747
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
V IY VL RRRG+V + GTP+++V AYLP ESFGF DLR++T GQA V
Sbjct 748 AAVGGIYGVLNRRRGHVFEESQVTGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCV 807
Query 122 FDNWAIVPGDPLD 134
FD+W ++PGDPL+
Sbjct 808 FDHWQVLPGDPLE 820
> Hs4503483
Length=858
Score = 165 bits (417), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 75/134 (55%), Positives = 95/134 (70%), Gaps = 0/134 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
EEN++GVRF + D + DA+HRGGGQIIP+ARR Y+++L A PRL+EPI EIQCP
Sbjct 694 EENMRGVRFDVHDVTLHADAIHRGGGQIIPTARRCLYASVLTAQPRLMEPIYLVEIQCPE 753
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
V IY VL R+RG+V + GTP+++V AYLP ESFGF DLR++T GQA V
Sbjct 754 QVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCV 813
Query 122 FDNWAIVPGDPLDK 135
FD+W I+PGDP D
Sbjct 814 FDHWQILPGDPFDN 827
> Hs22052810
Length=846
Score = 165 bits (417), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 75/134 (55%), Positives = 95/134 (70%), Gaps = 0/134 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
EEN++GVRF + D + DA+HRGGGQIIP+ARR Y+++L A PRL+EPI EIQCP
Sbjct 682 EENMRGVRFDVHDVTLHADAIHRGGGQIIPTARRCLYASVLTAQPRLMEPIYLVEIQCPE 741
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
V IY VL R+RG+V + GTP+++V AYLP ESFGF DLR++T GQA V
Sbjct 742 QVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCV 801
Query 122 FDNWAIVPGDPLDK 135
FD+W I+PGDP D
Sbjct 802 FDHWQILPGDPFDN 815
> 7302127
Length=832
Score = 158 bits (400), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 71/131 (54%), Positives = 92/131 (70%), Gaps = 0/131 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
+EN++GVRF + D + DA+HRGGGQIIP+ RR Y+A + A PRL+EP+ EIQCP
Sbjct 668 DENLRGVRFNIYDVTLHADAIHRGGGQIIPTTRRCLYAAAITAKPRLMEPVYLCEIQCPE 727
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
V IY VL RRRG+V + GTP+++V AYLP ESFGF DLR++T GQA V
Sbjct 728 VAVGGIYGVLNRRRGHVFEENQVVGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCV 787
Query 122 FDNWAIVPGDP 132
FD+W ++PGDP
Sbjct 788 FDHWQVLPGDP 798
> At1g56075
Length=855
Score = 156 bits (394), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 69/133 (51%), Positives = 96/133 (72%), Gaps = 0/133 (0%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPA 61
EEN++G+ F++ D + DA+HRGGGQ+IP+ARRV Y++ + A PRLLEP+ EIQ P
Sbjct 691 EENMRGICFEVCDVVLHSDAIHRGGGQVIPTARRVIYASQITAKPRLLEPVYMVEIQAPE 750
Query 62 DCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTV 121
+ IY+VL ++RG+V +M +PGTPLY + AYLP +ESFGF + LR TSGQA V
Sbjct 751 GALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFGFSSQLRAATSGQAFPQCV 810
Query 122 FDNWAIVPGDPLD 134
FD+W ++ DPL+
Sbjct 811 FDHWEMMSSDPLE 823
> YOR133w
Length=842
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 66/132 (50%), Positives = 87/132 (65%), Gaps = 0/132 (0%)
Query 3 ENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPAD 62
E ++ VR ++D + DA+HRGGGQIIP+ RR Y+ LLA P++ EP+ EIQCP
Sbjct 679 EEMRSVRVNILDVTLHADAIHRGGGQIIPTMRRATYAGFLLADPKIQEPVFLVEIQCPEQ 738
Query 63 CVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVF 122
V IY+VL ++RG V + +PGTPL+ V AYLP ESFGF +LR T GQA VF
Sbjct 739 AVGGIYSVLNKKRGQVVSEEQRPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVF 798
Query 123 DNWAIVPGDPLD 134
D+W+ + DPLD
Sbjct 799 DHWSTLGSDPLD 810
> YDR385w
Length=842
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 66/132 (50%), Positives = 87/132 (65%), Gaps = 0/132 (0%)
Query 3 ENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPAD 62
E ++ VR ++D + DA+HRGGGQIIP+ RR Y+ LLA P++ EP+ EIQCP
Sbjct 679 EEMRSVRVNILDVTLHADAIHRGGGQIIPTMRRATYAGFLLADPKIQEPVFLVEIQCPEQ 738
Query 63 CVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVF 122
V IY+VL ++RG V + +PGTPL+ V AYLP ESFGF +LR T GQA VF
Sbjct 739 AVGGIYSVLNKKRGQVVSEEQRPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVF 798
Query 123 DNWAIVPGDPLD 134
D+W+ + DPLD
Sbjct 799 DHWSTLGSDPLD 810
> SPCP31B10.07
Length=842
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 67/134 (50%), Positives = 93/134 (69%), Gaps = 0/134 (0%)
Query 1 IEENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCP 60
EEN++ RF ++D + DA+HRGGGQIIP+ARRV Y++ LLA+P + EP+ EIQ
Sbjct 677 FEENLRSCRFNILDVVLHADAIHRGGGQIIPTARRVVYASTLLASPIIQEPVFLVEIQVS 736
Query 61 ADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLT 120
+ + IY+VL ++RG+V + + GTPLY + AYLP ESFGF +LR T+GQA
Sbjct 737 ENAMGGIYSVLNKKRGHVFSEEQRVGTPLYNIKAYLPVNESFGFTGELRQATAGQAFPQL 796
Query 121 VFDNWAIVPGDPLD 134
VFD+W+ + GDPLD
Sbjct 797 VFDHWSPMSGDPLD 810
> SPAC513.01c
Length=812
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 67/134 (50%), Positives = 93/134 (69%), Gaps = 0/134 (0%)
Query 1 IEENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCP 60
EEN++ RF ++D + DA+HRGGGQIIP+ARRV Y++ LLA+P + EP+ EIQ
Sbjct 647 FEENLRSCRFNILDVVLHADAIHRGGGQIIPTARRVVYASTLLASPIIQEPVFLVEIQVS 706
Query 61 ADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLT 120
+ + IY+VL ++RG+V + + GTPLY + AYLP ESFGF +LR T+GQA
Sbjct 707 ENAMGGIYSVLNKKRGHVFSEEQRVGTPLYNIKAYLPVNESFGFTGELRQATAGQAFPQL 766
Query 121 VFDNWAIVPGDPLD 134
VFD+W+ + GDPLD
Sbjct 767 VFDHWSPMSGDPLD 780
> YKL173w
Length=1008
Score = 99.4 bits (246), Expect = 2e-21, Method: Composition-based stats.
Identities = 59/149 (39%), Positives = 81/149 (54%), Gaps = 4/149 (2%)
Query 2 EENVKGVRFKLVDAEIAKDA-VHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCP 60
EE + GV++KL+ + D + QIIP ++ Y LL A P LLEPI +I
Sbjct 810 EEPIYGVQYKLLSISVPSDVNIDVMKSQIIPLMKKACYVGLLTAIPILLEPIYEVDITVH 869
Query 61 ADCVSAIYTVLARRRGN-VARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCL 119
A + + ++ +RRG+ + + + GTPL V +P IES GFETDLR T+G MC
Sbjct 870 APLLPIVEELMKKRRGSRIYKTIKVAGTPLLEVRGQVPVIESAGFETDLRLSTNGLGMCQ 929
Query 120 TVFDN--WAIVPGDPLDKQIVFKPLEPAP 146
F + W VPGD LDK L+PAP
Sbjct 930 LYFWHKIWRKVPGDVLDKDAFIPKLKPAP 958
> ECU11g1460
Length=850
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 50/144 (34%), Positives = 78/144 (54%), Gaps = 1/144 (0%)
Query 1 IEENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCP 60
I E ++G++F+L DA + DA+HRG Q++ + + LL A P L EPI EI P
Sbjct 685 IGEVMRGLKFELKDAVLHADAIHRGINQLLQPVKNLCKGLLLAAGPILYEPIYEVEITTP 744
Query 61 ADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLT 120
D A+ T+L +RG PG ++ LP ESF F DL++ + G+A
Sbjct 745 NDYSGAVTTILLSKRGTAEDFKTLPGNDTTMITGTLPVKESFTFNEDLKSGSRGKAGASM 804
Query 121 VFDNWAIVPGDPLD-KQIVFKPLE 143
F +++I+PG+ D ++FK +E
Sbjct 805 RFSHYSILPGNLEDPNSLMFKTVE 828
> At3g22980
Length=1015
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 63/107 (58%), Gaps = 0/107 (0%)
Query 27 GQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPG 86
GQ++ + + +A+L PR++E + F E+ + + +Y VL+RRR + ++ + G
Sbjct 853 GQVMTAVKDACRAAVLQTNPRIVEAMYFCELNTAPEYLGPMYAVLSRRRARILKEEMQEG 912
Query 87 TPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWAIVPGDPL 133
+ L+ VHAY+P ESFGF +LR TSG A L V +W ++ DP
Sbjct 913 SSLFTVHAYVPVSESFGFADELRKGTSGGASALMVLSHWEMLEEDPF 959
> Hs13375760
Length=857
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 59/107 (55%), Gaps = 0/107 (0%)
Query 27 GQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPG 86
GQ+I + + AL + RL+ + +I D + +Y VL++R G V ++ K G
Sbjct 696 GQLIATMKEACRYALQVKPQRLMAAMYTCDIMATGDVLGRVYAVLSKREGRVLQEEMKEG 755
Query 87 TPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWAIVPGDPL 133
T ++I+ A LP ESFGF ++R TSG A VF +W I+P DP
Sbjct 756 TDMFIIKAVLPVAESFGFADEIRKRTSGLASPQLVFSHWEIIPSDPF 802
> YNL163c
Length=1110
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 57/108 (52%), Gaps = 0/108 (0%)
Query 26 GGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKP 85
G++I S R + A L +PR++ I +IQ D + +Y V+ +R G + + K
Sbjct 949 SGRLITSTRDAIHEAFLDWSPRIMWAIYSCDIQTSVDVLGKVYAVILQRHGKIISEEMKE 1008
Query 86 GTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWAIVPGDPL 133
GTP + + A++P +E+FG D+R TSG A VF + + DP
Sbjct 1009 GTPFFQIEAHVPVVEAFGLSEDIRKRTSGAAQPQLVFSGFECIDLDPF 1056
> SPCC553.08c
Length=1000
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 58/107 (54%), Gaps = 0/107 (0%)
Query 27 GQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPG 86
GQ+I + L +PRL+ + ++Q ++ + +Y V+++RRG V + K G
Sbjct 841 GQVISVVKESIRHGFLGWSPRLMLAMYSCDVQATSEVLGRVYGVVSKRRGRVIDEEMKEG 900
Query 87 TPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWAIVPGDPL 133
TP +IV A +P +ESFGF ++ TSG A +F + ++ +P
Sbjct 901 TPFFIVKALIPVVESFGFAVEILKRTSGAAYPQLIFHGFEMLDENPF 947
> At1g62750
Length=783
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 47/98 (47%), Gaps = 1/98 (1%)
Query 32 SARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPGTPLYI 91
+AR + A PR+LEPIM E+ P + + + L RRG + KPG L +
Sbjct 671 AARGAFREGMRKAGPRMLEPIMRVEVVTPEEHLGDVIGDLNSRRGQINSFGDKPGG-LKV 729
Query 92 VHAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWAIVP 129
V + +P E F + + LR T G+A + +VP
Sbjct 730 VDSLVPLAEMFQYVSTLRGMTKGRASYTMQLAKFDVVP 767
> Hs19923640
Length=779
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query 21 AVHRGGGQIIPSA--RRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNV 78
+H G + SA R AL A ++LEP+M E+ D +S + LA+RRGN+
Sbjct 656 TIHPGTSTTMISACVSRCVQKALKKADKQVLEPLMNLEVTVARDYLSPVLADLAQRRGNI 715
Query 79 ARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQA 116
+ + I ++P E G+ T LRT TSG A
Sbjct 716 QEIQTRQDNKVVI--GFVPLAEIMGYSTVLRTLTSGSA 751
> 7294103
Length=122
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 0/67 (0%)
Query 67 IYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWA 126
+Y V+ RR G + G+ + V LP IESF F ++R TSG A +F +W
Sbjct 1 MYAVIGRRHGKILSGDLTQGSGNFAVTCLLPVIESFNFAQEMRKQTSGLACPQLMFSHWE 60
Query 127 IVPGDPL 133
++ DP
Sbjct 61 VIDIDPF 67
> 7300890
Length=1093
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 34/111 (30%), Positives = 56/111 (50%), Gaps = 2/111 (1%)
Query 29 IIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPGTP 88
++ +A + L + RLLEPIM +I P++ +S I L+RRR + +PK G
Sbjct 578 VMATAAQCVQKLLSTSGTRLLEPIMALQIVAPSERISGIMADLSRRRALINDVLPK-GER 636
Query 89 LYIVHAYLPAIESFGFETDLRTHTSGQA-MCLTVFDNWAIVPGDPLDKQIV 138
++ P E G+ + LRT +SG A M + FD+ + + + Q V
Sbjct 637 NKMILVNAPLAELSGYSSALRTISSGTASMTMQPFDDRSWSANEDMKSQFV 687
> ECU09g0810
Length=615
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/78 (34%), Positives = 37/78 (47%), Gaps = 0/78 (0%)
Query 48 LLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETD 107
+LEP+ EI D + V++ G V P + L YLP ESFGFETD
Sbjct 513 VLEPLYLVEITHAKDAEDLVSEVISSSFGEVIHQSRFPFSTLESTLCYLPVPESFGFETD 572
Query 108 LRTHTSGQAMCLTVFDNW 125
LR ++ A C+ + W
Sbjct 573 LRVYSCSTADCVKMPLYW 590
> CE12082
Length=906
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 52/119 (43%), Gaps = 12/119 (10%)
Query 26 GGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPK- 84
GGQ++ + + +A RL+ + + + + ++ VL++R+ V + +
Sbjct 732 GGQMMTAIKASCSAAAKKLALRLVAAMYRCTVTTASQALGKVHAVLSQRKSKVGMENCRT 791
Query 85 -----------PGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWAIVPGDP 132
T L+ V + +P +ESF F LR TSG A F +W ++ DP
Sbjct 792 FSNIVLSEDINEATNLFEVVSLMPVVESFSFCDQLRKFTSGMASAQLQFSHWQVIDEDP 850
> 7297230
Length=729
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 56/120 (46%), Gaps = 20/120 (16%)
Query 5 VKGVRFKLVDAEIAKDAVHRGGGQIIPSARRV----AYSAL----LLATPRLLEPIMFSE 56
+ G+RF+L D GG I+ S+ A+ A+ + ++LEPIM E
Sbjct 582 LSGIRFRLQD----------GGHHIVDSSELAFMLAAHGAIKEVFQNGSWQILEPIMLVE 631
Query 57 IQCPADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQA 116
+ P + A+ L++R G + G + V+A +P + FG+ +LR+ T G+
Sbjct 632 VTAPEEFQGAVMGHLSKRHGIITGTEGTEG--WFTVYAEVPLNDMFGYAGELRSSTQGKG 689
> Hs18390331
Length=751
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 51/113 (45%), Gaps = 6/113 (5%)
Query 5 VKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCV 64
+ G+RF L D A V I + AL AT +LEPIM E+ P +
Sbjct 604 LSGLRFVLQDG--AHHMVDSNEISFIRAGEGALKQALANATLCILEPIMAVEVVAPNEFQ 661
Query 65 SAIYTVLARRRGNVARDMPKPGTPLYI-VHAYLPAIESFGFETDLRTHTSGQA 116
+ + RR G + + G Y ++A +P + FG+ T+LR+ T G+
Sbjct 662 GQVIAGINRRHGVIT---GQDGVEDYFTLYADVPLNDMFGYSTELRSCTEGKG 711
> Hs22043180
Length=485
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 51/113 (45%), Gaps = 6/113 (5%)
Query 5 VKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCPADCV 64
+ G+RF L D A V I + AL AT +LEPIM E+ P +
Sbjct 338 LSGLRFVLQDG--AHHMVDSNEISFIRAGEGALKQALANATLCILEPIMAVEVVAPNEFQ 395
Query 65 SAIYTVLARRRGNVARDMPKPGTPLYI-VHAYLPAIESFGFETDLRTHTSGQA 116
+ + RR G + + G Y ++A +P + FG+ T+LR+ T G+
Sbjct 396 GQVIAGINRRHGVIT---GQDGVEDYFTLYADVPLNDMFGYSTELRSCTEGKG 445
> At2g45030
Length=754
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 43/85 (50%), Gaps = 7/85 (8%)
Query 33 ARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRG-NVARDMPKPGTPLYI 91
A R+ Y+A A P +LEP+M E++ P + + + +R+G V D + +
Sbjct 645 AFRLCYTA---ARPVILEPVMLVELKVPTEFQGTVAGDINKRKGIIVGNDQEGDDS---V 698
Query 92 VHAYLPAIESFGFETDLRTHTSGQA 116
+ A +P FG+ T LR+ T G+
Sbjct 699 ITANVPLNNMFGYSTSLRSMTQGKG 723
> At1g45332
Length=754
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 43/85 (50%), Gaps = 7/85 (8%)
Query 33 ARRVAYSALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRG-NVARDMPKPGTPLYI 91
A R+ Y+A A P +LEP+M E++ P + + + +R+G V D + +
Sbjct 645 AFRLCYTA---ARPVILEPVMLVELKVPTEFQGTVAGDINKRKGIIVGNDQEGDDS---V 698
Query 92 VHAYLPAIESFGFETDLRTHTSGQA 116
+ A +P FG+ T LR+ T G+
Sbjct 699 ITANVPLNNMFGYSTSLRSMTQGKG 723
> At5g13650
Length=609
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/140 (24%), Positives = 58/140 (41%), Gaps = 15/140 (10%)
Query 6 KGVRFKLVDAEIAKDAVHRGGGQI-----IPSARRVAYSALL--------LATPRLLEPI 52
+ + K+ D E A + G G + I + RR Y ++ +LLEP
Sbjct 352 RNLAMKVEDGETADTFIVSGRGTLHITILIENMRREGYEFMVGPPKVINKRVNDKLLEPY 411
Query 53 MFSEIQCPADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAY-LPAIESFGFETDLRTH 111
+ ++ P + + +L +RRG + DM G+ Y +P G + T
Sbjct 412 EIATVEVPEAHMGPVVELLGKRRGQMF-DMQGVGSEGTTFLRYKIPTRGLLGLRNAILTA 470
Query 112 TSGQAMCLTVFDNWAIVPGD 131
+ G A+ TVFD++ GD
Sbjct 471 SRGTAILNTVFDSYGPWAGD 490
> CE19822
Length=750
Score = 35.8 bits (81), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 29/126 (23%), Positives = 52/126 (41%), Gaps = 4/126 (3%)
Query 1 IEENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPIMFSEIQCP 60
I+ + G+ ++ D + AV +I + + + + A LLEPIM E P
Sbjct 597 IKSRIAGIHVRIQDG--STHAVDSTEIAMINTMQNMMRESFEKANWLLLEPIMKVEATVP 654
Query 61 ADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLT 120
+ + T L +R + G I A P + FG+ ++LR+ T G+
Sbjct 655 TEFQGNVVTSLTQRNALITTTDSTEGYATVICEA--PLSDMFGYTSELRSLTEGKGEFSM 712
Query 121 VFDNWA 126
+ +A
Sbjct 713 EYSRYA 718
> SPBC1306.01c
Length=558
Score = 35.4 bits (80), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 34/78 (43%), Gaps = 2/78 (2%)
Query 39 SALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPA 98
+A L A P +LEPIM I P + + L +R+ + + + A +P
Sbjct 453 TAFLQANPMVLEPIMNVSITAPVEHQGGVIGNLDKRKATIVD--SDTDEDEFTLQAEVPL 510
Query 99 IESFGFETDLRTHTSGQA 116
F + +D+R T G+
Sbjct 511 NSMFSYSSDIRALTKGKG 528
> YLR069c
Length=761
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 2/87 (2%)
Query 40 ALLLATPRLLEPIMFSEIQCPADCVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAI 99
A L A P ++EPIM + P + + +L + + V +D + G + + A
Sbjct 656 AFLRAQPVIMEPIMNVSVTSPNEFQGNVIGLLNKLQA-VIQD-TENGHDEFTLKAECALS 713
Query 100 ESFGFETDLRTHTSGQAMCLTVFDNWA 126
FGF T LR T G+ F ++A
Sbjct 714 TMFGFATSLRASTQGKGEFSLEFSHYA 740
> Hs18591704
Length=851
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 32/134 (23%), Positives = 57/134 (42%), Gaps = 15/134 (11%)
Query 11 KLVDAEIAKDAVHRG-GGQIIPSARRVAYSALLLAT--PRLLEPIMFS-----EIQCPAD 62
++ + E+A+D R ++P ++ Y+ L T P+LLE + CP D
Sbjct 655 EIAEDELARDWTKRRMKDDLVPETSQLNYTRPGLPTLNPQLLEAWKNEVKEKGHVNCPND 714
Query 63 CVSAIYTVLARRRGNVARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSGQAMCLTVF 122
C AIY+ ++ + ++A G L + L + F E+ ++ H + T
Sbjct 715 CCEAIYSSVSGLKAHLASCS--KGAHLAGKYRCLLCPKEFSSESGVKYH-----ILKTHA 767
Query 123 DNWAIVPGDPLDKQ 136
+NW DP K
Sbjct 768 ENWFRTSADPPPKH 781
> CE27459
Length=689
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 40/96 (41%), Gaps = 6/96 (6%)
Query 25 GGGQIIPS-----ARRVAYSALLLATPRLLEPIMFSEIQCPADC-VSAIYTVLARRRGNV 78
GG+I P+ A++ AL L EP+M +I+ D I L RRR +
Sbjct 564 SGGKINPALLSACAQKCVSEALSSGEMVLTEPVMEVQIEIRNDDPTQPILNELLRRRAHF 623
Query 79 ARDMPKPGTPLYIVHAYLPAIESFGFETDLRTHTSG 114
T + + A LP E+ +RT TSG
Sbjct 624 EHSDATESTEIRRICAILPLSETENLSKTVRTLTSG 659
> CE15447
Length=305
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 3/51 (5%)
Query 2 EENVKGVRFKLVDAEIAKDAVHRGGGQIIPSARRVAYSALLLATPRLLEPI 52
E V+ ++F L K G II SA+R+AY A + P + +P+
Sbjct 108 ENGVQTLKFDLTSGAGVK---RSAGDPIIDSAKRIAYGAQSVVEPEIPQPM 155
> At4g35140
Length=493
Score = 28.1 bits (61), Expect = 5.8, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 93 HAYLPAIESFGFETDLRTHTSGQAMCLTVFDNWAIVP 129
H ++P + S G E+D++ TS A T+ +N ++P
Sbjct 337 HPHIPVLASSGIESDIKVWTSKAAERATLPENIELLP 373
Lambda K H
0.323 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40