bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5446_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
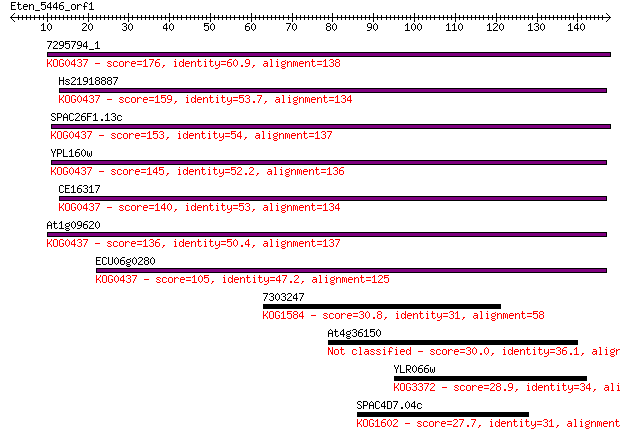
7295794_1 176 2e-44
Hs21918887 159 2e-39
SPAC26F1.13c 153 1e-37
YPL160w 145 2e-35
CE16317 140 7e-34
At1g09620 136 1e-32
ECU06g0280 105 4e-23
7303247 30.8 0.99
At4g36150 30.0 1.7
YLR066w 28.9 3.2
SPAC4D7.04c 27.7 8.6
> 7295794_1
Length=1143
Score = 176 bits (446), Expect = 2e-44, Method: Composition-based stats.
Identities = 84/143 (58%), Positives = 99/143 (69%), Gaps = 9/143 (6%)
Query 10 HSPQTMHQ-----FSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAY 64
H +T H+ F L+WL E+ACSR+YGLGT LPW D LIESLSDSTIYMA+
Sbjct 492 HGMETFHEEARNNFEACLNWLHEYACSRTYGLGTKLPW----DDKWLIESLSDSTIYMAF 547
Query 65 YTVAHLMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEF 124
YTV HL+QGG G+ PGP G+ P+D+T + +DYIF PK T + E+L LR EF
Sbjct 548 YTVVHLLQGGTFRGEKPGPFGIKPSDMTGEIWDYIFFKETPLPKKTAIKQEHLAVLRREF 607
Query 125 SYWYPMDLRVSGKDLIFNHLTFC 147
YWYPMDLRVSGKDLI NHLTFC
Sbjct 608 EYWYPMDLRVSGKDLIQNHLTFC 630
> Hs21918887
Length=1176
Score = 159 bits (402), Expect = 2e-39, Method: Composition-based stats.
Identities = 72/134 (53%), Positives = 99/134 (73%), Gaps = 5/134 (3%)
Query 13 QTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQ 72
+T F L WL+E ACSR+YGLGT LPW D LIESLSDSTIYMA+YTVAHL+Q
Sbjct 556 ETRRNFEATLGWLQEHACSRTYGLGTHLPW----DEQWLIESLSDSTIYMAFYTVAHLLQ 611
Query 73 GGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDL 132
GG+++G PLG+ P +T++ +DY+F ++ P T+++ E L++L+ EF +WYP+DL
Sbjct 612 GGNLHGQAESPLGIRPQQMTKEVWDYVF-FKEAPFPKTQIAKEKLDQLKQEFEFWYPVDL 670
Query 133 RVSGKDLIFNHLTF 146
RVSGKDL+ NHL++
Sbjct 671 RVSGKDLVPNHLSY 684
> SPAC26F1.13c
Length=1111
Score = 153 bits (386), Expect = 1e-37, Method: Composition-based stats.
Identities = 74/138 (53%), Positives = 94/138 (68%), Gaps = 8/138 (5%)
Query 11 SPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHL 70
SP+ + F L WL +WAC+RSYGLGT LPW D L+ESL+DSTIYMAYYT+ HL
Sbjct 578 SPEVRNGFLKTLDWLSQWACARSYGLGTRLPW----DPQFLVESLTDSTIYMAYYTICHL 633
Query 71 MQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEP-PKDTKVSLENLNRLRAEFSYWYP 129
+ D+YG PG L + P +T + +D++F R P PK+T +S E L RL EF Y+YP
Sbjct 634 LHS-DVYGKVPGALNIKPEQMTPEVWDHVF--RQAPKPKNTSISDEALARLCREFQYFYP 690
Query 130 MDLRVSGKDLIFNHLTFC 147
D+R SGKDL+ NHLTFC
Sbjct 691 FDIRASGKDLVPNHLTFC 708
> YPL160w
Length=1090
Score = 145 bits (366), Expect = 2e-35, Method: Composition-based stats.
Identities = 71/136 (52%), Positives = 95/136 (69%), Gaps = 6/136 (4%)
Query 11 SPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHL 70
+P+ + F VL WLK WA R+YGLGT LPW D L+ESLSDSTIY ++YT+AHL
Sbjct 570 APEVKNAFEGVLDWLKNWAVCRTYGLGTRLPW----DEKYLVESLSDSTIYQSFYTIAHL 625
Query 71 MQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPM 130
+ D YG+ GPLG++ +T++ FDYIF +D+ K+T + L L +LR EF Y+YP+
Sbjct 626 L-FKDYYGNEIGPLGISADQMTDEVFDYIFQHQDDV-KNTNIPLPALQKLRREFEYFYPL 683
Query 131 DLRVSGKDLIFNHLTF 146
D+ +SGKDLI NHLTF
Sbjct 684 DVSISGKDLIPNHLTF 699
> CE16317
Length=1186
Score = 140 bits (354), Expect = 7e-34, Method: Composition-based stats.
Identities = 71/134 (52%), Positives = 83/134 (61%), Gaps = 4/134 (2%)
Query 13 QTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQ 72
+T + WL E+ACSRSYGLGT LPW D LIESLSDSTIY AYYTVAHL+Q
Sbjct 557 ETRRGLETTVDWLHEYACSRSYGLGTKLPW----DTQYLIESLSDSTIYNAYYTVAHLLQ 612
Query 73 GGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDL 132
G G GP G+ +T+ + Y+FL K V E L LR EF YWYP+D+
Sbjct 613 QGAFDGSVVGPAGIKADQMTDASWSYVFLGEIYDSKTMPVEEEKLKSLRKEFMYWYPIDM 672
Query 133 RVSGKDLIFNHLTF 146
R SGKDLI NHLT+
Sbjct 673 RASGKDLIGNHLTY 686
> At1g09620
Length=1084
Score = 136 bits (343), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 69/138 (50%), Positives = 93/138 (67%), Gaps = 9/138 (6%)
Query 10 HSPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAH 69
+S +T H F LSWL +WACSRS+GLGT +PW D L+ESLSDS++YMAYYTVAH
Sbjct 556 YSDETRHGFEHTLSWLNQWACSRSFGLGTRIPW----DEQFLVESLSDSSLYMAYYTVAH 611
Query 70 LMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEP-PKDTKVSLENLNRLRAEFSYWY 128
+ GD+Y + + P + ++ ++Y+F D P PK + + L+ ++ EF YWY
Sbjct 612 IFHDGDMYKGSKSLI--RPQQMNDEVWEYLFC--DGPYPKSSDIPSAVLSEMKQEFDYWY 667
Query 129 PMDLRVSGKDLIFNHLTF 146
P+DLRVSGKDLI NHLTF
Sbjct 668 PLDLRVSGKDLIQNHLTF 685
> ECU06g0280
Length=874
Score = 105 bits (261), Expect = 4e-23, Method: Composition-based stats.
Identities = 59/126 (46%), Positives = 78/126 (61%), Gaps = 11/126 (8%)
Query 22 LSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQGGDIYGDTP 81
L W+ +W SRS+GLGT +PW D+ LI+SLSDSTIYMA YT H + Y D
Sbjct 399 LEWIGKWGFSRSFGLGTRIPW----DSEYLIDSLSDSTIYMAMYTFKHFL-----YRDLE 449
Query 82 GPLGLTPAD-LTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDLRVSGKDLI 140
G L P++ L++ ++YIFL R +D E L+ R F+Y+YP+DLRV GKDL+
Sbjct 450 GKDELFPSNRLSDDVWNYIFLNR-SITEDLAPYEEILSNCRESFNYFYPIDLRVGGKDLL 508
Query 141 FNHLTF 146
NHL F
Sbjct 509 KNHLIF 514
> 7303247
Length=346
Score = 30.8 bits (68), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 63 AYYTVAHLMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRL 120
Y ++ + + + T P L P + E+ I+++RD PKD VS +LNRL
Sbjct 141 GYEALSEIPRSQRRFIKTHFPFSLMPPSVLEKKCKVIYVVRD--PKDVAVSYYHLNRL 196
> At4g36150
Length=1179
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 3/61 (4%)
Query 79 DTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDLRVSGKD 138
DT L LT D E F + F + PP T NL+RL A+++ P+ L++ GK+
Sbjct 363 DTYEVLRLTGRDSFEYFSYFAFSGKLCPPVRT---FMNLSRLFADYAKGNPLALKILGKE 419
Query 139 L 139
L
Sbjct 420 L 420
> YLR066w
Length=184
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 7/47 (14%)
Query 95 FFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDLRVSGKDLIF 141
F+D I +D+ D +N LR+++S W D + GKDL+F
Sbjct 113 FWDKIIKSKDDAVID-------VNDLRSKYSIWDIEDGKFEGKDLVF 152
> SPAC4D7.04c
Length=264
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 1/42 (2%)
Query 86 LTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYW 127
+TP D+ E F+ L++D P D + + RL ++F W
Sbjct 187 ITPEDIDEDIFEKNLLIKDSLPLDLLIRTSGVERL-SDFMLW 227
Lambda K H
0.322 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40