bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5488_orf1
Length=123
Score E
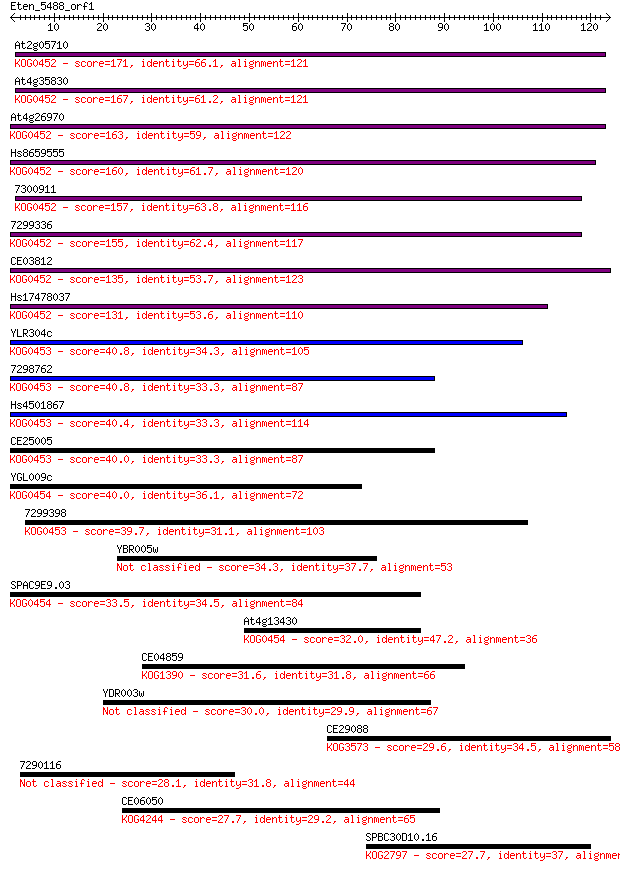
Sequences producing significant alignments: (Bits) Value
At2g05710 171 3e-43
At4g35830 167 6e-42
At4g26970 163 6e-41
Hs8659555 160 4e-40
7300911 157 5e-39
7299336 155 2e-38
CE03812 135 3e-32
Hs17478037 131 3e-31
YLR304c 40.8 6e-04
7298762 40.8 7e-04
Hs4501867 40.4 8e-04
CE25005 40.0 0.001
YGL009c 40.0 0.001
7299398 39.7 0.001
YBR005w 34.3 0.061
SPAC9E9.03 33.5 0.11
At4g13430 32.0 0.32
CE04859 31.6 0.37
YDR003w 30.0 0.97
CE29088 29.6 1.4
7290116 28.1 4.3
CE06050 27.7 5.0
SPBC30D10.16 27.7 5.2
> At2g05710
Length=898
Score = 171 bits (433), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 80/121 (66%), Positives = 95/121 (78%), Gaps = 0/121 (0%)
Query 2 KAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRIH 61
K+GL + L E GF + GYGC TCIGN+GE + V AITE D+V A+VLSGNRNFEGR+H
Sbjct 488 KSGLQEYLNEQGFNIVGYGCTTCIGNSGEINESVGAAITENDIVAAAVLSGNRNFEGRVH 547
Query 62 PLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPVIEQC 121
PLTRA YLASPPLVVAYALAG ++IDF+TEPIG GK VFLRDIWPT +EI V++
Sbjct 548 PLTRANYLASPPLVVAYALAGTVNIDFETEPIGKGKNGKDVFLRDIWPTTEEIAEVVQSS 607
Query 122 L 122
+
Sbjct 608 V 608
> At4g35830
Length=898
Score = 167 bits (422), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 74/121 (61%), Positives = 93/121 (76%), Gaps = 0/121 (0%)
Query 2 KAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRIH 61
K+GL K L +LGF + GYGC TCIGN+G+ V AI + DLV ++VLSGNRNFEGR+H
Sbjct 488 KSGLQKYLNQLGFSIVGYGCTTCIGNSGDIHEAVASAIVDNDLVASAVLSGNRNFEGRVH 547
Query 62 PLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPVIEQC 121
PLTRA YLASPPLVVAYALAG +DIDF+T+PIG +GK +F RDIWP+ E+ V++
Sbjct 548 PLTRANYLASPPLVVAYALAGTVDIDFETQPIGTGKDGKQIFFRDIWPSNKEVAEVVQSS 607
Query 122 L 122
+
Sbjct 608 V 608
> At4g26970
Length=907
Score = 163 bits (413), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 72/122 (59%), Positives = 96/122 (78%), Gaps = 0/122 (0%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRI 60
+++GL ++L + GF + GYGC TCIGN+G DPEV AI D++ A+VLSGNRNFEGR+
Sbjct 496 DRSGLRESLTKQGFEIVGYGCTTCIGNSGNLDPEVASAIEGTDIIPAAVLSGNRNFEGRV 555
Query 61 HPLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPVIEQ 120
HP TRA YLASPPLVVAYALAG +DIDF+ EPIG ++GK V+LRD+WP+ +E+ V++
Sbjct 556 HPQTRANYLASPPLVVAYALAGTVDIDFEKEPIGTRSDGKSVYLRDVWPSNEEVAQVVQY 615
Query 121 CL 122
+
Sbjct 616 SV 617
> Hs8659555
Length=889
Score = 160 bits (406), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 74/120 (61%), Positives = 94/120 (78%), Gaps = 0/120 (0%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRI 60
+++G+ L +LGF + GYGCMTCIGN+G V +AIT+GDLV VLSGNRNFEGR+
Sbjct 483 QESGVMPYLSQLGFDVVGYGCMTCIGNSGPLPEPVVEAITQGDLVAVGVLSGNRNFEGRV 542
Query 61 HPLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPVIEQ 120
HP TRA YLASPPLV+AYA+AG + IDF+ EP+G + +G+ VFL+DIWPTRDEI+ V Q
Sbjct 543 HPNTRANYLASPPLVIAYAIAGTIRIDFEKEPLGVNAKGQQVFLKDIWPTRDEIQAVERQ 602
> 7300911
Length=902
Score = 157 bits (397), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 74/116 (63%), Positives = 89/116 (76%), Gaps = 0/116 (0%)
Query 2 KAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRIH 61
++G+ L++LGF + GYGCMTCIGN+G D V I + LV VLSGNRNFEGRIH
Sbjct 497 ESGVIPYLEQLGFDIVGYGCMTCIGNSGPLDENVVNTIEKNGLVCCGVLSGNRNFEGRIH 556
Query 62 PLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPV 117
P TRA YLASP LV+AYA+AGR+DIDF+ EP+G D+ GK VFLRDIWPTR EI+ V
Sbjct 557 PNTRANYLASPLLVIAYAIAGRVDIDFEIEPLGVDSNGKEVFLRDIWPTRSEIQEV 612
> 7299336
Length=899
Score = 155 bits (391), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 73/117 (62%), Positives = 90/117 (76%), Gaps = 0/117 (0%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRI 60
+++G+ L++LGF + GYGCMTCIGN+G + V I + LV A VLSGNRNFEGRI
Sbjct 493 KESGVIPYLEKLGFDIVGYGCMTCIGNSGPLEENVVNTIEKNGLVCAGVLSGNRNFEGRI 552
Query 61 HPLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPV 117
HP TRA YLASP LV+AYA+AGR+DIDF+ EP+G D GK VFL+DIWPTR EI+ V
Sbjct 553 HPNTRANYLASPLLVIAYAIAGRVDIDFEKEPLGVDANGKNVFLQDIWPTRSEIQEV 609
> CE03812
Length=887
Score = 135 bits (339), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 66/123 (53%), Positives = 87/123 (70%), Gaps = 1/123 (0%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRI 60
E +GL L+++GF +AGYGCMTCIGN+G D VT+AI E +LVVA VLSGNRNFEGRI
Sbjct 482 EASGLLPYLEKIGFNIAGYGCMTCIGNSGPLDEPVTKAIEENNLVVAGVLSGNRNFEGRI 541
Query 61 HPLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPVIEQ 120
HP RA YLASPPL V Y++ G +++D + + +GK + L DIWPTR E+ E+
Sbjct 542 HPHVRANYLASPPLAVLYSIIGNVNVDINGV-LAVTPDGKEIRLADIWPTRKEVAKFEEE 600
Query 121 CLR 123
++
Sbjct 601 FVK 603
> Hs17478037
Length=963
Score = 131 bits (329), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 59/110 (53%), Positives = 76/110 (69%), Gaps = 0/110 (0%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRI 60
+G+ L +LGF + GYGC C+GNT V A+ +GDLV +LSGN+NFEGR+
Sbjct 558 SSSGVLPYLSKLGFEIVGYGCSICVGNTAPLSDAVLNAVKQGDLVTCGILSGNKNFEGRL 617
Query 61 HPLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPT 110
RA YLASPPLVVAYA+AG ++IDF TEP+G D GK ++L DIWP+
Sbjct 618 CDCVRANYLASPPLVVAYAIAGTVNIDFQTEPLGTDPTGKNIYLHDIWPS 667
> YLR304c
Length=778
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 51/107 (47%), Gaps = 11/107 (10%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRI 60
E+ G + KE G + C CIG D I +GD V S NRNF R
Sbjct 425 ERDGQLETFKEFGGIVLANACGPCIGQWDRRD------IKKGDKNTI-VSSYNRNFTSRN 477
Query 61 --HPLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLR 105
+P T A ++ASP LV A+A+AG + + T+ + D +G L+
Sbjct 478 DGNPQTHA-FVASPELVTAFAIAGDLRFNPLTDKL-KDKDGNEFMLK 522
> 7298762
Length=683
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 43/89 (48%), Gaps = 10/89 (11%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGR- 59
E+ G+ + + G + C CIG D + +GD V S NRNF GR
Sbjct 332 ERDGISEVFDKFGGTVLANACGPCIGQWDRKD------VKKGDKNTI-VTSYNRNFTGRN 384
Query 60 -IHPLTRAAYLASPPLVVAYALAGRMDID 87
+P T ++ SP LV A ++AGR+D +
Sbjct 385 DANPATHC-FVTSPELVTALSIAGRLDFN 412
> Hs4501867
Length=780
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 57/116 (49%), Gaps = 13/116 (11%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGR- 59
E+ G + L++LG + C CIG D I +G+ V S NRNF GR
Sbjct 428 ERDGYAQILRDLGGIVLANACGPCIGQWDRKD------IKKGEKNTI-VTSYNRNFTGRN 480
Query 60 -IHPLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEI 114
+P T A ++ SP +V A A+AG + + +T+ + T+GK L P DE+
Sbjct 481 DANPETHA-FVTSPEIVTALAIAGTLKFNPETDYL-TGTDGKKFRLEA--PDADEL 532
> CE25005
Length=777
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 45/89 (50%), Gaps = 10/89 (11%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGR- 59
E+ GL K + G + C CIG ++D Q + +G+ V S NRNF GR
Sbjct 425 ERDGLSKIFADFGGMVLANACGPCIG---QWD---RQDVKKGEKNTI-VTSYNRNFTGRN 477
Query 60 -IHPLTRAAYLASPPLVVAYALAGRMDID 87
+P T ++ SP + A A++GR+D +
Sbjct 478 DANPATHG-FVTSPDITTAMAISGRLDFN 505
> YGL009c
Length=779
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 34/72 (47%), Gaps = 11/72 (15%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRI 60
E GLDK +E GF GC C+G +P++ A + NRNFEGR
Sbjct 401 EAEGLDKIFQEAGFEWREAGCSICLG----MNPDILDAYER------CASTSNRNFEGRQ 450
Query 61 HPLTRAAYLASP 72
L+R +L SP
Sbjct 451 GALSR-THLMSP 461
> 7299398
Length=769
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 51/105 (48%), Gaps = 11/105 (10%)
Query 4 GLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGR--IH 61
G+ L++ G + C CIG D ++ + T V S NRNF GR +
Sbjct 415 GIIDVLEKFGGTVLANACGPCIGQWDRKDVKMGEKNT-------IVTSYNRNFTGRNDAN 467
Query 62 PLTRAAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRD 106
P T ++ SP + A A+AGR+D + T+ + T+GK L++
Sbjct 468 PATHC-FVTSPEMATALAIAGRLDFNPMTDEL-TGTDGKTFKLKE 510
> YBR005w
Length=213
Score = 34.3 bits (77), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 26/53 (49%), Gaps = 7/53 (13%)
Query 23 TCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRIHPLTRAAYLASPPLV 75
T T ++ PE T+ E DL G + G HP +AAY+A PPLV
Sbjct 95 TVQQRTDDYVPEYTETANEHDL-------GYYDQRGEFHPNDKAAYVAPPPLV 140
> SPAC9E9.03
Length=758
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 36/85 (42%), Gaps = 13/85 (15%)
Query 1 EKAGLDKALKELGFYLAGYGCMTCIG-NTGEFDPEVTQAITEGDLVVASVLSGNRNFEGR 59
E GLD+ E GF GC C+G N + P A T NRNFEGR
Sbjct 400 EAEGLDQIFIEAGFDWREAGCSMCLGMNPDQLKPYERCASTS-----------NRNFEGR 448
Query 60 IHPLTRAAYLASPPLVVAYALAGRM 84
R +L SP + A A+ G +
Sbjct 449 QGAKGR-THLVSPAMAAAAAIKGHL 472
> At4g13430
Length=509
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 49 VLSGNRNFEGRIHPLTRAAYLASPPLVVAYALAGRM 84
V + NRNF GR+ YLASP A AL GR+
Sbjct 466 VSTTNRNFPGRMGHKEGQIYLASPYTAAASALTGRV 501
> CE04859
Length=390
Score = 31.6 bits (70), Expect = 0.37, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 29/66 (43%), Gaps = 11/66 (16%)
Query 28 TGEFDPEVTQAITEGDLVVASVLSGNRNFEGRIHPLTRAAYLASPPLVVAYALAGRMDID 87
TG P ++ +G VA+V+ G HPL +VA+A AGR ID
Sbjct 239 TGTITPANASSLNDG--AVATVVVGENALPQGAHPLAE---------LVAFAEAGRAPID 287
Query 88 FDTEPI 93
F P+
Sbjct 288 FTVAPV 293
> YDR003w
Length=210
Score = 30.0 bits (66), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 28/67 (41%), Gaps = 10/67 (14%)
Query 20 GCMTCIGNTGEFDPEVTQAITEGDLVVASVLSGNRNFEGRIHPLTRAAYLASPPLVVAYA 79
G C+ ++ PE T+ E DL G + G HP + YLA PPL A
Sbjct 93 GTQRCVE---DYVPEYTETANENDL-------GFYDERGEFHPNGKTEYLAPPPLSEEQA 142
Query 80 LAGRMDI 86
+ D+
Sbjct 143 SSTDKDL 149
> CE29088
Length=503
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 9/66 (13%)
Query 66 AAYLASPPLVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDE--------IRPV 117
A LA+ P +V + A R + + P+ + +G P FLR W RD +RP
Sbjct 343 APRLANKPKIV-FVQACRGERRDNGFPVLDSVDGVPAFLRRGWDNRDGPLFNFLGCVRPQ 401
Query 118 IEQCLR 123
++Q R
Sbjct 402 VQQVWR 407
> 7290116
Length=319
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 3 AGLDKALKELGFYLAGYGCMTCIGNTGEFDPEVTQAITEGDLVV 46
+ L K++++ G +LA C++ + N E E+ +A +GDL V
Sbjct 83 SALSKSIEDAGIHLAE--CLSGLANMTEIQAEIEEASPKGDLDV 124
> CE06050
Length=277
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 31/68 (45%), Gaps = 3/68 (4%)
Query 24 CIGNTGEFDPEVTQAITEGDL-VVASVLSGNRNFEGRIHPLTRAAY--LASPPLVVAYAL 80
C+G G+F+P+ + DL V+ + GN F +I P + LA+ + L
Sbjct 189 CVGAIGDFEPQELDELLHRDLKVIQDSIKGNFLFGDKITPADATVFGQLATVYYPIRSHL 248
Query 81 AGRMDIDF 88
+D DF
Sbjct 249 TDVLDKDF 256
> SPBC30D10.16
Length=287
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 74 LVVAYALAGRMDIDFDTEPIGNDTEGKPVFLRDIWPTRDEIRPVIE 119
L V AL+ R +D+ PI N T G + D+ RD+I+ V E
Sbjct 51 LAVLEALSSR-QVDYAVLPIENSTNGAVIPAYDLLKGRDDIQAVGE 95
Lambda K H
0.320 0.141 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191192512
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40