bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
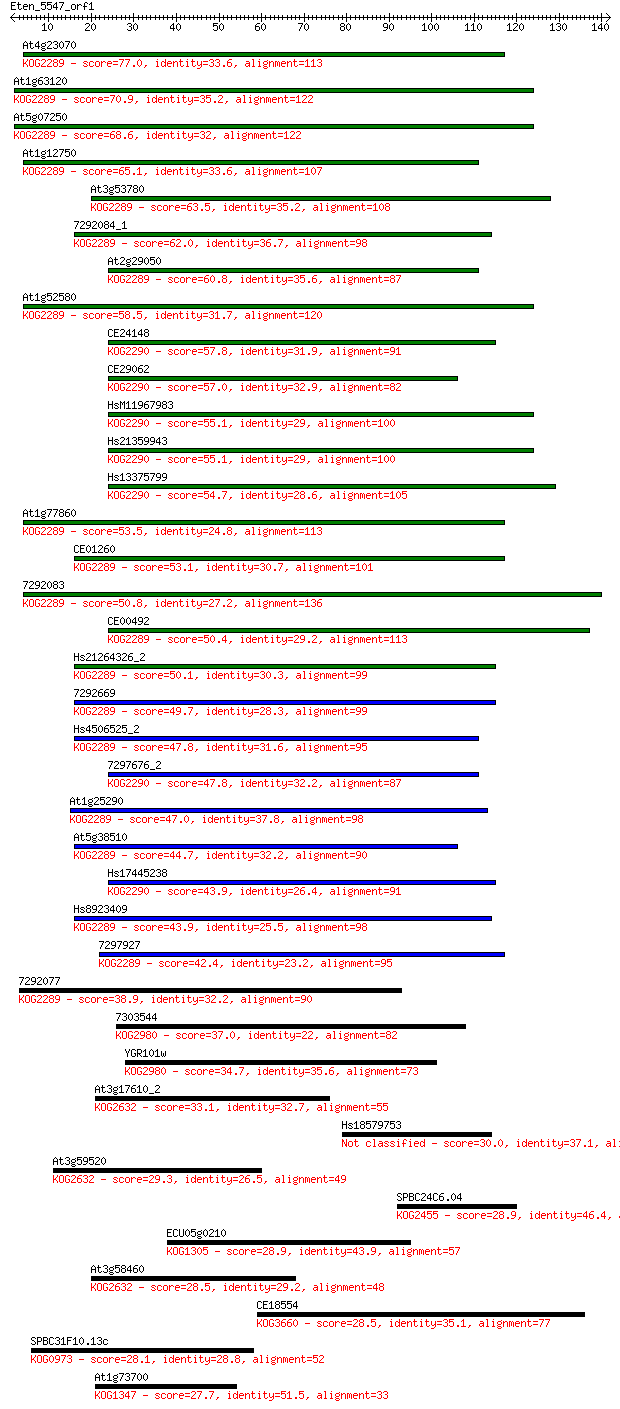
Query= Eten_5547_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
At4g23070 77.0 1e-14
At1g63120 70.9 7e-13
At5g07250 68.6 4e-12
At1g12750 65.1 4e-11
At3g53780 63.5 1e-10
7292084_1 62.0 3e-10
At2g29050 60.8 9e-10
At1g52580 58.5 4e-09
CE24148 57.8 7e-09
CE29062 57.0 1e-08
HsM11967983 55.1 5e-08
Hs21359943 55.1 5e-08
Hs13375799 54.7 5e-08
At1g77860 53.5 1e-07
CE01260 53.1 2e-07
7292083 50.8 7e-07
CE00492 50.4 1e-06
Hs21264326_2 50.1 1e-06
7292669 49.7 2e-06
Hs4506525_2 47.8 6e-06
7297676_2 47.8 7e-06
At1g25290 47.0 1e-05
At5g38510 44.7 6e-05
Hs17445238 43.9 1e-04
Hs8923409 43.9 1e-04
7297927 42.4 3e-04
7292077 38.9 0.003
7303544 37.0 0.011
YGR101w 34.7 0.059
At3g17610_2 33.1 0.18
Hs18579753 30.0 1.4
At3g59520 29.3 2.5
SPBC24C6.04 28.9 3.2
ECU05g0210 28.9 3.5
At3g58460 28.5 4.0
CE18554 28.5 4.1
SPBC31F10.13c 28.1 6.1
At1g73700 27.7 7.8
> At4g23070
Length=313
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 38/114 (33%), Positives = 69/114 (60%), Gaps = 1/114 (0%)
Query 4 PTERVLYQLGA-TYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRT 62
P+ L ++GA +G + Q WRL+ ++LH G++HLL N+ + ++G+ E ++G
Sbjct 82 PSSSTLEKMGALAWGKIVHKRQVWRLLTCMWLHAGVIHLLANMCCVAYIGVRLEQQFGFV 141
Query 63 NFLILYFSSALIGNMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLDER 116
+Y S G++ + L +++VGAS+A FGL+G++L+E+LI W D +
Sbjct 142 RVGTIYLVSGFCGSILSCLFLEDAISVGASSALFGLLGAMLSELLINWTTYDNK 195
> At1g63120
Length=317
Score = 70.9 bits (172), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 72/123 (58%), Gaps = 1/123 (0%)
Query 2 FEPTERVLYQLGATYGPGIRHL-QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYG 60
F P+ L ++GA + H Q WRL+ ++LH GI+HLL N+L ++ +G+ E ++G
Sbjct 86 FGPSSSTLEKMGALEWRKVVHEHQGWRLLSCMWLHAGIIHLLTNMLSLIFIGIRLEQQFG 145
Query 61 RTNFLILYFSSALIGNMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNM 120
++Y S L G++ + L S++VGAS A FGL+G++L+E+L W + +
Sbjct 146 FIRVGLIYLISGLGGSILSSLFLQESISVGASGALFGLLGAMLSELLTNWTIYANKAAAL 205
Query 121 YTL 123
TL
Sbjct 206 ITL 208
> At5g07250
Length=346
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 70/123 (56%), Gaps = 1/123 (0%)
Query 2 FEPTERVLYQLGA-TYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYG 60
F P+ L +LGA + + + WRL+ ++LH G++HL N+L ++ +G+ E ++G
Sbjct 110 FGPSSHTLEKLGALEWSKVVEKKEGWRLLTCIWLHAGVIHLGANMLSLVFIGIRLEQQFG 169
Query 61 RTNFLILYFSSALIGNMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNM 120
++Y S + G++ + L S++VGAS A FGL+GS+L+E+ W + +
Sbjct 170 FVRIGVIYLLSGIGGSVLSSLFIRNSISVGASGALFGLLGSMLSELFTNWTIYSNKIAAL 229
Query 121 YTL 123
TL
Sbjct 230 LTL 232
> At1g12750
Length=307
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 64/108 (59%), Gaps = 1/108 (0%)
Query 4 PTERVLYQLGA-TYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRT 62
P+ L +LGA + ++ + WRLI ++LH GI+HL+ N+ ++ G+ E ++G
Sbjct 75 PSSSTLEKLGALDWKKVVQGNEKWRLITAMWLHAGIIHLVMNMFDVIIFGIRLEQQFGFI 134
Query 63 NFLILYFSSALIGNMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVW 110
++Y S G++ + L S++VGAS A GL+G++L+E+L W
Sbjct 135 RIGLIYLISGFGGSILSALFLQKSISVGASGALLGLMGAMLSELLTNW 182
> At3g53780
Length=361
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 38/108 (35%), Positives = 63/108 (58%), Gaps = 5/108 (4%)
Query 20 IRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFT 79
++ + WRL+ +LH G+VHLL N+L +L +G+ E++ G +LY S G++ +
Sbjct 106 VKGDEGWRLLSCNWLHGGVVHLLMNMLTLLFIGIRMELRIG-----LLYLISGFGGSILS 160
Query 80 VLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNMYTLDMTV 127
L +++VGAS A FGL+G +L+EI I W + + TL + V
Sbjct 161 ALFLRSNISVGASGAVFGLLGGMLSEIFINWTIYSNKVVTIVTLVLIV 208
> 7292084_1
Length=356
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 54/98 (55%), Gaps = 1/98 (1%)
Query 16 YGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIG 75
Y P RH + WR + + LH G +HL FNV L GL E+ +G T +YFS L G
Sbjct 105 YRPDKRH-EIWRFLFYMVLHAGWLHLGFNVAVQLVFGLPLEMVHGSTRIACIYFSGVLAG 163
Query 76 NMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKL 113
++ T + P VGAS + L+ + LA +L+ +H++
Sbjct 164 SLGTSIFDPDVFLVGASGGVYALLAAHLANVLLNYHQM 201
> At2g29050
Length=372
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 54/87 (62%), Gaps = 0/87 (0%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
+ WRL ++LH G+ H+L N+L ++ +G+ E ++G +LY S G++ + L
Sbjct 154 EVWRLFTCIWLHAGVFHVLANMLSLIFIGIRLEQEFGFVRIGLLYMISGFGGSLLSSLFN 213
Query 84 PCSLAVGASTAGFGLVGSILAEILIVW 110
++VGAS A FGL+G++L+E+L W
Sbjct 214 RAGISVGASGALFGLLGAMLSELLTNW 240
> At1g52580
Length=309
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 68/122 (55%), Gaps = 2/122 (1%)
Query 4 PTERVLYQLGATYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTN 63
P+ L +LGA + + WRLI ++LH G +HL+ N++ ++ +G+ E ++G
Sbjct 85 PSIPTLRKLGALERRLVEEGERWRLISCIWLHGGFLHLMANMISLMCIGMRLEQEFGFMR 144
Query 64 FLILYFSSALIGNMFTVLMRPCS--LAVGASTAGFGLVGSILAEILIVWHKLDERTRNMY 121
LY S L G++ + L ++VGAS A FGL+G++L+E++ W + + +
Sbjct 145 IGALYVISGLGGSLVSCLTDSQGERVSVGASGALFGLLGAMLSELITNWTIYENKCTALM 204
Query 122 TL 123
TL
Sbjct 205 TL 206
> CE24148
Length=851
Score = 57.8 bits (138), Expect = 7e-09, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 49/91 (53%), Gaps = 0/91 (0%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
Q +RL +F+H G++HL ++ F ++ +E G ILYF+S + GN+ + +
Sbjct 574 QIYRLFTSLFIHAGVIHLALSMAFQMYFMAYQENLIGSKRMAILYFASGISGNLASAIFV 633
Query 84 PCSLAVGASTAGFGLVGSILAEILIVWHKLD 114
P VG S+A G+ S++ E+ H LD
Sbjct 634 PYYPTVGPSSAQCGVFSSVVVELWHFRHLLD 664
> CE29062
Length=727
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 48/82 (58%), Gaps = 0/82 (0%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
Q +RL +F+H G++HL ++LF ++ D E ILYF+S + GN+ + +
Sbjct 451 QFYRLFTSLFVHAGVIHLALSLLFQYYVMKDLENLIASKRMAILYFASGIGGNLASAIFV 510
Query 84 PCSLAVGASTAGFGLVGSILAE 105
P + AVG S+A G++ +++ E
Sbjct 511 PYNPAVGPSSAQCGILAAVIVE 532
> HsM11967983
Length=855
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 49/100 (49%), Gaps = 0/100 (0%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
Q +RL + +FLH GI+H L ++ F + + D E G I+Y S + GN+ + +
Sbjct 652 QFYRLWLSLFLHAGILHCLVSICFQMTVLRDLEKLAGWHRIAIIYLLSGVTGNLASAIFL 711
Query 84 PCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNMYTL 123
P VG + + FG++ + E+ W L R + L
Sbjct 712 PYRAEVGPAGSQFGILACLFVELFQSWQILARPWRAFFKL 751
> Hs21359943
Length=855
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 49/100 (49%), Gaps = 0/100 (0%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
Q +RL + +FLH GI+H L ++ F + + D E G I+Y S + GN+ + +
Sbjct 652 QFYRLWLSLFLHAGILHCLVSICFQMTVLRDLEKLAGWHRIAIIYLLSGVTGNLASAIFL 711
Query 84 PCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNMYTL 123
P VG + + FG++ + E+ W L R + L
Sbjct 712 PYRAEVGPAGSQFGILACLFVELFQSWQILARPWRAFFKL 751
> Hs13375799
Length=619
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 30/105 (28%), Positives = 51/105 (48%), Gaps = 0/105 (0%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
Q +RL + +FLH G+VH L +V+F + + D E G I++ S + GN+ + +
Sbjct 416 QFYRLWLSLFLHAGVVHCLVSVVFQMTILRDLEKLAGWHRIAIIFILSGITGNLASAIFL 475
Query 84 PCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNMYTLDMTVF 128
P VG + + FGL+ + E+ W L+ + L V
Sbjct 476 PYRAEVGPAGSQFGLLACLFVELFQSWPLLERPWKAFLNLSAIVL 520
> At1g77860
Length=319
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 62/114 (54%), Gaps = 1/114 (0%)
Query 4 PTERVLYQLGA-TYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRT 62
P+ L +G ++ + + WR++ +LH G+ HL N+ ++ +G+ E ++G
Sbjct 100 PSASTLEHMGGLSWKALTENHEIWRILTSPWLHSGLFHLFINLGSLIFVGIYMEQQFGPL 159
Query 63 NFLILYFSSALIGNMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLDER 116
++YF S ++G++F VL ++ + A FGL+G++L+ + W+ + +
Sbjct 160 RIAVIYFLSGIMGSLFAVLFVRNIPSISSGAAFFGLIGAMLSALAKNWNLYNSK 213
> CE01260
Length=419
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 55/101 (54%), Gaps = 1/101 (0%)
Query 16 YGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIG 75
+ P +R +AWR +FLH G+ HLL NV+ L +G+ E+ + +Y + G
Sbjct 221 FAPKLRG-EAWRFTSYMFLHAGLNHLLGNVIIQLLVGIPLEVAHKIWRIGPIYLLAVTSG 279
Query 76 NMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLDER 116
++ + P SL VGAS + L+ + +A +++ WH++ R
Sbjct 280 SLLQYAIDPNSLLVGASAGVYALIFAHVANVILNWHEMPLR 320
> 7292083
Length=355
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 37/136 (27%), Positives = 64/136 (47%), Gaps = 14/136 (10%)
Query 4 PTERVLYQLGATYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTN 63
P++ VL Y P R LQ WR +FLH HL FN++ L G+ E+ +G
Sbjct 135 PSDSVL-----VYRPD-RRLQVWRFFSYMFLHANWFHLGFNIVIQLFFGIPLEVMHGTAR 188
Query 64 FLILYFSSALIGNMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNMYTL 123
++Y + G++ T ++ VGAS + L+ + LA I + + + +
Sbjct 189 IGVIYMAGVFAGSLGTSVVDSEVFLVGASGGVYALLAAHLANITLNYAHMKSAS------ 242
Query 124 DMTVFGALMILLSYDI 139
T G+++I +S D+
Sbjct 243 --TQLGSVVIFVSCDL 256
> CE00492
Length=397
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 58/113 (51%), Gaps = 3/113 (2%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
+ WRL ++VGI H++FN+L L +G+ E+ + R ILYF L G++ ++ +
Sbjct 130 ELWRLFTYCLINVGIFHIIFNILIQLAIGVPLELVH-RWRIYILYFMGVLFGSILSLALD 188
Query 84 PCSLAVGASTAGFGLVGSILAEILIVWHKLDERTRNMYTLDMTVFGALMILLS 136
P G + F L+ S + I + +++ T + L VF AL +L+
Sbjct 189 PTVFLCGGAAGSFSLIASHITTIATNFKEMENATCRLPIL--IVFAALDYVLA 239
> Hs21264326_2
Length=302
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 1/99 (1%)
Query 16 YGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIG 75
Y P +R Q WR + +F+H GI HL NV+ L +G+ E+ +G T ++Y + + G
Sbjct 101 YHPQLR-AQVWRYLTYIFMHAGIEHLGLNVVLQLLVGVPLEMVHGATRIGLVYVAGVVAG 159
Query 76 NMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLD 114
++ + + VG+S + LV + LA I++ W +
Sbjct 160 SLAVSVADMTAPVVGSSGGVYALVSAHLANIVMNWSGMK 198
> 7292669
Length=219
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 56/99 (56%), Gaps = 1/99 (1%)
Query 16 YGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIG 75
Y P R+ + WR + +F+HVGI+HL+ N++ + +G+ E+ + ++Y + L G
Sbjct 26 YNPYKRY-EGWRFVSYMFVHVGIMHLMMNLIIQIFLGIALELVHHWWRVGLVYLAGVLAG 84
Query 76 NMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKLD 114
+M T L P GAS + L+ + +A I++ + +++
Sbjct 85 SMGTSLTSPRIFLAGASGGVYALITAHIATIIMNYSEME 123
> Hs4506525_2
Length=302
Score = 47.8 bits (112), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 50/95 (52%), Gaps = 1/95 (1%)
Query 16 YGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIG 75
Y PG R +AWR + +F+HVG+ L FN L L +G+ E+ +G +LY + L G
Sbjct 101 YHPGHR-ARAWRFLTYMFMHVGLEQLGFNALLQLMIGVPLEMVHGLLRISLLYLAGVLAG 159
Query 76 NMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVW 110
++ + + VG S + L + LA +++ W
Sbjct 160 SLTVSITDMRAPVVGGSGGVYALCSAHLANVVMNW 194
> 7297676_2
Length=689
Score = 47.8 bits (112), Expect = 7e-06, Method: Composition-based stats.
Identities = 28/87 (32%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
Q +RL+ + +H GI+HL ++F D E G I+Y S GN+ + ++
Sbjct 352 QLYRLLTSLCMHAGILHLAITLIFQHLFLADLERLIGTVRTAIVYIMSGFAGNLTSAILV 411
Query 84 PCSLAVGASTAGFGLVGSILAEILIVW 110
P VG S + G+V S++A L+VW
Sbjct 412 PHRPEVGPSASLSGVVASLIA--LLVW 436
> At1g25290
Length=369
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/108 (34%), Positives = 51/108 (47%), Gaps = 12/108 (11%)
Query 15 TYGPGIRHL----QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFS 70
T+G I L Q WRL LH +HL+ N + +G E G FL +Y +
Sbjct 179 TWGAKINSLIERGQLWRLATASVLHANPMHLMINCYSLNSIGPTAESLGGPKRFLAVYLT 238
Query 71 SALIGNMFTVLMRPCSL------AVGASTAGFGLVGSILAEILIVWHK 112
SA+ + VL S +VGAS A FGLVGS+ + ++ HK
Sbjct 239 SAVAKPILRVLGSAMSYWFNKAPSVGASGAIFGLVGSV--AVFVIRHK 284
> At5g38510
Length=434
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 43/94 (45%), Gaps = 5/94 (5%)
Query 16 YGPGIRHL----QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSS 71
YG I L + WRL+ P+FLH GI H+ + +L G YG F ++Y
Sbjct 218 YGAKINDLILAGEWWRLVTPMFLHSGIPHVALSSWALLTFGPKVCRDYGLFTFCLIYILG 277
Query 72 ALIGNMFTVLMRPCSLAVGASTAGFGLVGSILAE 105
+ GN F + VG + F L+G+ L +
Sbjct 278 GVSGN-FMSFLHTADPTVGGTGPAFALIGAWLVD 310
> Hs17445238
Length=619
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 45/91 (49%), Gaps = 0/91 (0%)
Query 24 QAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMR 83
Q +RL + +FLH I+H L ++ F + + D E G + I+Y S + GN+ + +
Sbjct 428 QFYRLWLSLFLHTRILHYLVSICFQMTIPRDLEKLAGWHHIAIIYLLSGVTGNLASAIFL 487
Query 84 PCSLAVGASTAGFGLVGSILAEILIVWHKLD 114
V + + FG++ + E+ W L+
Sbjct 488 LYRAEVVPAGSQFGILACLFMELFQSWQILE 518
> Hs8923409
Length=292
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 50/98 (51%), Gaps = 1/98 (1%)
Query 16 YGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIG 75
Y P R +AWR I + +H G+ H+L N+ L +G+ E+ + ++Y + + G
Sbjct 101 YSPEKRE-EAWRFISYMLVHAGVQHILGNLCMQLVLGIPLEMVHKGLRVGLVYLAGVIAG 159
Query 76 NMFTVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKL 113
++ + + P VGAS + L+G +L+ + ++
Sbjct 160 SLASSIFDPLRYLVGASGGVYALMGGYFMNVLVNFQEM 197
> 7297927
Length=263
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 49/95 (51%), Gaps = 0/95 (0%)
Query 22 HLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVL 81
+++ WRL+ + LH HL N+ F +G+ E++ G ++Y + G++
Sbjct 104 NVEYWRLLTYMLLHSDYWHLSLNICFQCFIGICLEVEQGHWRLAVVYMVGGVAGSLANAW 163
Query 82 MRPCSLAVGASTAGFGLVGSILAEILIVWHKLDER 116
++P +GAS + ++GS + +++ + +L R
Sbjct 164 LQPHLHLMGASAGVYAMLGSHVPHLVLNFSQLSHR 198
> 7292077
Length=235
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 43/90 (47%), Gaps = 5/90 (5%)
Query 3 EPTERVLYQLGATYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRT 62
+P E L Y P R LQ WR + LH +HL +NVL L G+ E+ +G
Sbjct 12 DPPEDSLL----VYRPDQR-LQLWRFLSYALLHASWLHLGYNVLTQLLFGVPLELVHGSL 66
Query 63 NFLILYFSSALIGNMFTVLMRPCSLAVGAS 92
++Y + L G++ T ++ VGAS
Sbjct 67 RTGVIYMAGVLAGSLGTSVVDSEVFLVGAS 96
> 7303544
Length=351
Score = 37.0 bits (84), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/82 (21%), Positives = 37/82 (45%), Gaps = 0/82 (0%)
Query 26 WRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLMRPC 85
W + + F H +HL N+ + + G+ FL +Y S+ + ++ +VL +
Sbjct 186 WPMFLSTFSHYSAMHLFANMYVMHSFANAAAVSLGKEQFLAVYLSAGVFSSLMSVLYKAA 245
Query 86 SLAVGASTAGFGLVGSILAEIL 107
+ G S G + ++LA +
Sbjct 246 TSQAGMSLGASGAIMTLLAYVC 267
> YGR101w
Length=346
Score = 34.7 bits (78), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 40/79 (50%), Gaps = 6/79 (7%)
Query 28 LIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIGNMFTVLM-RPCS 86
+I F H HL N+L + G G +NF LY +SA+ G++F++ +
Sbjct 186 IIGSAFSHQEFWHLGMNMLALWSFGTSLATMLGASNFFSLYMNSAIAGSLFSLWYPKLAR 245
Query 87 LAV-----GASTAGFGLVG 100
LA+ GAS A FG++G
Sbjct 246 LAIVGPSLGASGALFGVLG 264
> At3g17610_2
Length=302
Score = 33.1 bits (74), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Query 21 RHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLILYFSSALIG 75
+H RL + F HV HL++N++ +L G+ E G + F + F+ LIG
Sbjct 40 KHKDLKRLFLSAFYHVNEPHLVYNMMSLLWKGIKLETSMGSSEFASMVFT--LIG 92
> Hs18579753
Length=168
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 79 TVLMRPCSLAVGASTAGFGLVGSILAEILIVWHKL 113
TV M+P A+ +++ G G++G IL + LI+ L
Sbjct 133 TVAMQPMPCALASNSPGLGVLGPILTQTLILQESL 167
> At3g59520
Length=269
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 11 QLGATYGPGIRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKY 59
Q+G +Y I WR+I H+ ++HL+FN+ + +G+ +++ +
Sbjct 38 QVGLSYETAIEG-HYWRMITSALSHISVLHLVFNMSALWSLGVVEQLGH 85
> SPBC24C6.04
Length=548
Score = 28.9 bits (63), Expect = 3.2, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 92 STAGFGLVGSILAEILIVWHKLDERTRN 119
+T +GL GSI A+ +V KL +R RN
Sbjct 457 TTTPYGLTGSIFAQDRVVVRKLTDRLRN 484
> ECU05g0210
Length=426
Score = 28.9 bits (63), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 33/61 (54%), Gaps = 5/61 (8%)
Query 38 IVHLLFNVLFILHMGLDKEIKYGRTNFLILY-FSSALIG---NMFTVLMRPCSLAVGAST 93
IV + FNVLFI LD EI+ + IL+ SAL+G +F+ R LAV AS
Sbjct 119 IVDVFFNVLFIYFPFLD-EIRKSMAVYKILFGLGSALVGLTLYVFSTFSRKMFLAVSASA 177
Query 94 A 94
+
Sbjct 178 S 178
> At3g58460
Length=411
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 20 IRHLQAWRLIMPVFLHVGIVHLLFNVLFILHMGLDKEIKYGRTNFLIL 67
I Q +R + H ++H+LFN++ ++ MG + E G L L
Sbjct 59 ISRFQVYRFYTAIIFHGSLLHVLFNMMALVPMGSELERIMGSVRLLYL 106
> CE18554
Length=706
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 27/88 (30%), Positives = 36/88 (40%), Gaps = 17/88 (19%)
Query 59 YGRTNFLILYFSSALIGNMFTVLMRPCSLAVGASTAGFGL----VGSILAEILIVW---- 110
YGR N L A + MF C+ +G + FG V +L ILIV
Sbjct 520 YGRRNQL------ADMKEMFGESKNKCTSWIGQHSPYFGFNWMFVTPVLGSILIVLASIR 573
Query 111 ---HKLDERTRNMYTLDMTVFGALMILL 135
++ DE NMY L + G + LL
Sbjct 574 QYPYQTDENKPNMYPLAFDIVGWAVCLL 601
> SPBC31F10.13c
Length=932
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 27/55 (49%), Gaps = 3/55 (5%)
Query 6 ERVLYQLGATYGPGIRHLQAWRLIMPVFLHVGIVHLL---FNVLFILHMGLDKEI 57
E + LG+T+G G +H + WR + H + L ++ ++ +GLD I
Sbjct 104 EEAIPGLGSTFGSGEKHTENWRSYRRLLGHDNDIQDLCWSYDSQLVVSVGLDSSI 158
> At1g73700
Length=476
Score = 27.7 bits (60), Expect = 7.8, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 21 RHLQAWRLIMPVFLHVGI---VHLLFNVLFILHMGL 53
R LQA + PVF+ GI +HLL LF+L GL
Sbjct 166 RFLQAQNNVFPVFVCSGITTCLHLLLCWLFVLKTGL 201
Lambda K H
0.332 0.146 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40