bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5573_orf2
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
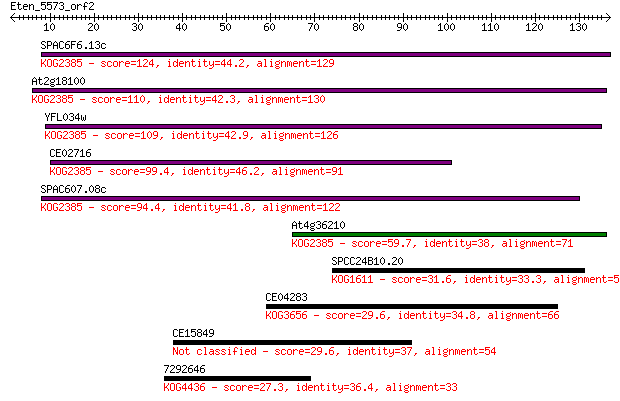
SPAC6F6.13c 124 4e-29
At2g18100 110 6e-25
YFL034w 109 1e-24
CE02716 99.4 2e-21
SPAC607.08c 94.4 6e-20
At4g36210 59.7 2e-09
SPCC24B10.20 31.6 0.43
CE04283 29.6 1.8
CE15849 29.6 1.9
7292646 27.3 7.4
> SPAC6F6.13c
Length=778
Score = 124 bits (312), Expect = 4e-29, Method: Composition-based stats.
Identities = 57/130 (43%), Positives = 83/130 (63%), Gaps = 1/130 (0%)
Query 8 AVGQRPVTLAGYSMGARVIFYCLQALWKKRKFHCVHDVVLMGLPASMNAAQWRQARQVTS 67
++G RPVTL GYS+GARVI+YCL+ L KK++F + +V L G P W +A V S
Sbjct 564 SLGVRPVTLVGYSLGARVIYYCLRELEKKKEFSIIENVYLFGTPVIFKRTSWLKAASVVS 623
Query 68 GRLVNVYCRTDWLLAFLYRWMEFRL-QVAGLAPVTSVPGVENVDVTGLVKSHANYPEKVP 126
GR VN Y + DW+L +L+R + +VAGL + +PG+EN+DVT LV H Y E +P
Sbjct 624 GRFVNGYKKNDWILGYLFRATSGGIGRVAGLRQIDCIPGIENIDVTNLVSGHLAYRESMP 683
Query 127 QIMSFISIEM 136
+++ + E+
Sbjct 684 ILLAAVGFEV 693
> At2g18100
Length=655
Score = 110 bits (276), Expect = 6e-25, Method: Composition-based stats.
Identities = 55/131 (41%), Positives = 79/131 (60%), Gaps = 2/131 (1%)
Query 6 KAAVGQRPVTLAGYSMGARVIFYCLQALWKKRK-FHCVHDVVLMGLPASMNAAQWRQARQ 64
K G RPVTL G+S+GARV+F CLQ L + K V VVL+G P S+ WR R+
Sbjct 516 KGLQGNRPVTLVGFSLGARVVFKCLQTLAETEKNAEIVERVVLLGAPISIKNENWRDVRK 575
Query 65 VTSGRLVNVYCRTDWLLAFLYRWMEFRLQVAGLAPVTSVPGVENVDVTGLVKSHANYPEK 124
+ +GR +NVY DW L +R +AG+ PV +PG+E+VDVT +V+ H++Y K
Sbjct 576 MVAGRFINVYATNDWTLGVAFRASLLSQGLAGIQPV-CIPGIEDVDVTDMVEGHSSYLWK 634
Query 125 VPQIMSFISIE 135
QI+ + ++
Sbjct 635 TQQILERLELD 645
> YFL034w
Length=1073
Score = 109 bits (272), Expect = 1e-24, Method: Composition-based stats.
Identities = 54/127 (42%), Positives = 75/127 (59%), Gaps = 1/127 (0%)
Query 9 VGQRPVTLAGYSMGARVIFYCLQALWKKRKFHCVHDVVLMGLPASMNAAQWRQARQVTSG 68
+G RP+TL G+S+GARVIF CL L KK+ + +V L G PA M Q AR V SG
Sbjct 748 LGARPITLVGFSIGARVIFSCLIELCKKKALGLIENVYLFGTPAVMKKEQLVMARSVVSG 807
Query 69 RLVNVYCRTDWLLAFLYRWMEFRLQ-VAGLAPVTSVPGVENVDVTGLVKSHANYPEKVPQ 127
R VN Y DW LA+L+R V G++ + +V G+EN++ T V H NY + +P+
Sbjct 808 RFVNGYSDKDWFLAYLFRAAAGGFSAVMGISTIENVEGIENINCTEFVDGHLNYRKSMPK 867
Query 128 IMSFISI 134
++ I I
Sbjct 868 LLKRIGI 874
> CE02716
Length=531
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 42/92 (45%), Positives = 63/92 (68%), Gaps = 1/92 (1%)
Query 10 GQRPVTLAGYSMGARVIFYCLQALWKKRK-FHCVHDVVLMGLPASMNAAQWRQARQVTSG 68
G+RP+TL G+S+GARVIF+CL + K+ + + DV+L+G P + + +W + V SG
Sbjct 421 GKRPITLIGFSLGARVIFHCLLTMSKRSESVGIIEDVILLGAPVTASPKEWSKVCTVVSG 480
Query 69 RLVNVYCRTDWLLAFLYRWMEFRLQVAGLAPV 100
R++N YC TDWLL FLYR M + ++AG P+
Sbjct 481 RVINGYCETDWLLRFLYRTMSAQFRIAGTGPI 512
> SPAC607.08c
Length=579
Score = 94.4 bits (233), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 51/123 (41%), Positives = 72/123 (58%), Gaps = 1/123 (0%)
Query 8 AVGQRPVTLAGYSMGARVIFYCLQALWKKRKFHCVHDVVLMGLPASMNAAQWRQARQVTS 67
A G RPVTL G+S+GAR I CL L + + + V +V++MG P +A W + R V +
Sbjct 385 AQGMRPVTLIGFSLGARTILECLLHLADRGETNLVENVIVMGAPMPTDAKLWLKMRCVVA 444
Query 68 GRLVNVYCRTDWLLAFLYRWMEFRLQVAGLAPVT-SVPGVENVDVTGLVKSHANYPEKVP 126
GR VNVY +D++L +YR + AGL PV+ +ENVDV LV+ H Y V
Sbjct 445 GRFVNVYSASDYVLQLVYRVNSAQSTAAGLGPVSLDSNTLENVDVGDLVEGHLQYRWLVA 504
Query 127 QIM 129
+I+
Sbjct 505 KIL 507
> At4g36210
Length=516
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 27/71 (38%), Positives = 43/71 (60%), Gaps = 1/71 (1%)
Query 65 VTSGRLVNVYCRTDWLLAFLYRWMEFRLQVAGLAPVTSVPGVENVDVTGLVKSHANYPEK 124
+ +GR +NVY DW L +R +AG+ P+ +PG+ENVDVT +V+ H++Y K
Sbjct 437 MVAGRFINVYATNDWTLGIAFRASLISQGLAGIQPIC-IPGIENVDVTDMVEGHSSYLWK 495
Query 125 VPQIMSFISIE 135
QI+ + I+
Sbjct 496 TQQILERLEID 506
> SPCC24B10.20
Length=254
Score = 31.6 bits (70), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 2/59 (3%)
Query 74 YCRTDWLLAFLYRWMEFRLQVAGLAPVTSVPGVENVD--VTGLVKSHANYPEKVPQIMS 130
Y ++ L + + + F L+ G V+ PGV N D V + K + YPE V I S
Sbjct 159 YGQSKAALNYTMKEISFELEKDGFVVVSIHPGVVNTDMFVNAMQKLASKYPEMVESIKS 217
> CE04283
Length=490
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 9/67 (13%)
Query 59 WRQARQVTSGRLVNVYCRTDWLLAFLYRWMEFRLQVAGLAPVT-SVPGVENVDVTGLVKS 117
WRQ R V++GRL ++ L++ LQVA L V V + NV +T +K+
Sbjct 216 WRQCRLVSAGRLPDITQSIQLLMSI--------LQVAFLYIVPLFVLSIFNVKLTRFLKT 267
Query 118 HANYPEK 124
+AN K
Sbjct 268 NANKMSK 274
> CE15849
Length=376
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 38 KFHCVHDVVLMGLPASMNAAQWRQARQVTSGRLVNVYCRT--DWLLAFLYRWMEFR 91
K C+ V L PAS + AQ + +G LVN Y R D+++A L + EF+
Sbjct 230 KGTCIQPVTL---PASWSIAQASCHHKGNNGHLVNEYSREKHDFVMAILKTYKEFQ 282
> 7292646
Length=803
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 36 KRKFHCVHDVVLMGLPASMNAAQWRQARQVTSG 68
KRK CV ++V G+P A W+Q + G
Sbjct 103 KRKNPCVSELVRRGIPHHFRAIVWQQLSGASDG 135
Lambda K H
0.325 0.135 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1425342594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40