bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5606_orf3
Length=81
Score E
Sequences producing significant alignments: (Bits) Value
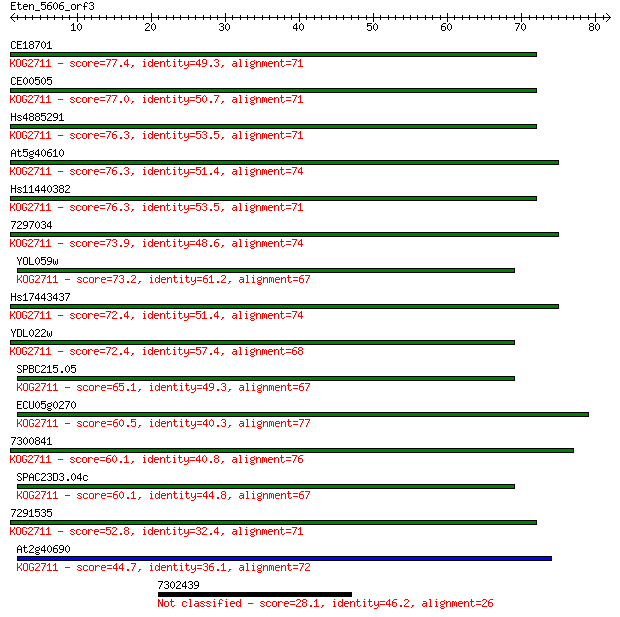
CE18701 77.4 6e-15
CE00505 77.0 7e-15
Hs4885291 76.3 1e-14
At5g40610 76.3 1e-14
Hs11440382 76.3 1e-14
7297034 73.9 6e-14
YOL059w 73.2 1e-13
Hs17443437 72.4 2e-13
YDL022w 72.4 2e-13
SPBC215.05 65.1 3e-11
ECU05g0270 60.5 8e-10
7300841 60.1 1e-09
SPAC23D3.04c 60.1 1e-09
7291535 52.8 2e-07
At2g40690 44.7 5e-05
7302439 28.1 3.9
> CE18701
Length=374
Score = 77.4 bits (189), Expect = 6e-15, Method: Composition-based stats.
Identities = 35/71 (49%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGALKN+VA A GF +G+G N K+AI+RLG+ E K F+ F+ + T ++S G AD
Sbjct 226 CGALKNIVACAAGFADGLGWAYNVKSAIIRLGLLETKKFVEHFYPSSVGHTYFESCGVAD 285
Query 61 VITTVFGGNCR 71
+ITT +GG R
Sbjct 286 LITTCYGGRNR 296
> CE00505
Length=351
Score = 77.0 bits (188), Expect = 7e-15, Method: Composition-based stats.
Identities = 36/71 (50%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGALKNVVA A GF +G+G G NTKAA++RLG+ E F+ ++ T ++S G AD
Sbjct 203 CGALKNVVACAAGFTDGLGYGDNTKAAVIRLGLMETTKFVEHYYPGSNLQTFFESCGIAD 262
Query 61 VITTVFGGNCR 71
+ITT +GG R
Sbjct 263 LITTCYGGRNR 273
> Hs4885291
Length=349
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 38/72 (52%), Positives = 50/72 (69%), Gaps = 1/72 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTF-FGDLLADTLYDSAGYA 59
CGALKNVVA+ GFC+G+G G NTKAA++RLG+ E+ F F G + + T +S G A
Sbjct 200 CGALKNVVAVGAGFCDGLGFGDNTKAAVIRLGLMEMIAFAKLFCSGPVSSATFLESCGVA 259
Query 60 DVITTVFGGNCR 71
D+ITT +GG R
Sbjct 260 DLITTCYGGRNR 271
> At5g40610
Length=400
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 38/74 (51%), Positives = 48/74 (64%), Gaps = 2/74 (2%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CG LKNVVA+A GF +G+ G NTKAAI+R+G+ E+K F + T ++S G AD
Sbjct 253 CGTLKNVVAIAAGFVDGLEMGNNTKAAIMRIGLREMKALSKLLFPSVKDSTFFESCGVAD 312
Query 61 VITTVFGGNCRARR 74
VITT GG R RR
Sbjct 313 VITTCLGG--RNRR 324
> Hs11440382
Length=349
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 38/72 (52%), Positives = 50/72 (69%), Gaps = 1/72 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTF-FGDLLADTLYDSAGYA 59
CGALKNVVA+ GFC+G+G G NTKAA++RLG+ E+ F F G + + T +S G A
Sbjct 200 CGALKNVVAVGAGFCDGLGFGDNTKAAVIRLGLMEMIAFAKLFCSGPVSSATFLESCGVA 259
Query 60 DVITTVFGGNCR 71
D+ITT +GG R
Sbjct 260 DLITTCYGGRNR 271
> 7297034
Length=360
Score = 73.9 bits (180), Expect = 6e-14, Method: Composition-based stats.
Identities = 36/74 (48%), Positives = 49/74 (66%), Gaps = 2/74 (2%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGALKN+VA GF +G+ G NTKAA++RLG+ E+ F+ F+ T ++S G AD
Sbjct 202 CGALKNIVACGAGFVDGLKLGDNTKAAVIRLGLMEMIRFVDVFYPGSKLSTFFESCGVAD 261
Query 61 VITTVFGGNCRARR 74
+ITT +GG R RR
Sbjct 262 LITTCYGG--RNRR 273
> YOL059w
Length=440
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 41/68 (60%), Positives = 46/68 (67%), Gaps = 1/68 (1%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLY-DSAGYAD 60
GALKNVVALA GF EGMG G N AAI RLG+ EI F FF + +T Y +SAG AD
Sbjct 291 GALKNVVALACGFVEGMGWGNNASAAIQRLGLGEIIKFGRMFFPESKVETYYQESAGVAD 350
Query 61 VITTVFGG 68
+ITT GG
Sbjct 351 LITTCSGG 358
> Hs17443437
Length=351
Score = 72.4 bits (176), Expect = 2e-13, Method: Composition-based stats.
Identities = 38/75 (50%), Positives = 50/75 (66%), Gaps = 3/75 (4%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFF-GDLLADTLYDSAGYA 59
CGALKN+VA+ GFC+G+ G NTKAA++RLG+ E+ F F G + T +S G A
Sbjct 202 CGALKNIVAVGAGFCDGLRCGDNTKAAVIRLGLMEMIAFARIFCKGQVSTATFLESCGVA 261
Query 60 DVITTVFGGNCRARR 74
D+ITT +GG R RR
Sbjct 262 DLITTCYGG--RNRR 274
> YDL022w
Length=391
Score = 72.4 bits (176), Expect = 2e-13, Method: Composition-based stats.
Identities = 39/69 (56%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLY-DSAGYA 59
CGALKNVVAL GF EG+G G N AAI R+G+ EI F FF + +T Y +SAG A
Sbjct 241 CGALKNVVALGCGFVEGLGWGNNASAAIQRVGLGEIIRFGQMFFPESREETYYQESAGVA 300
Query 60 DVITTVFGG 68
D+ITT GG
Sbjct 301 DLITTCAGG 309
> SPBC215.05
Length=385
Score = 65.1 bits (157), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/67 (49%), Positives = 44/67 (65%), Gaps = 0/67 (0%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
GALKNVVA+A GF +G+ G NTKAAI+R G+ E++ F TFF + S G AD+
Sbjct 229 GALKNVVAMAVGFADGLEWGGNTKAAIMRRGLLEMQKFATTFFDSDPRTMVEQSCGIADL 288
Query 62 ITTVFGG 68
+T+ GG
Sbjct 289 VTSCLGG 295
> ECU05g0270
Length=345
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 47/83 (56%), Gaps = 6/83 (7%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
G LKN+V++A GF EG+G TNTK AI R G E++ F F+ ++L+ S+G D+
Sbjct 199 GTLKNIVSMAYGFAEGLGYCTNTKVAIFRNGFAEMRKFFKFFYPMATTESLFQSSGVGDL 258
Query 62 ITTVFGG------NCRARRRLNL 78
+ + G A +R+NL
Sbjct 259 LVSCMSGRNFGCARLMAEKRMNL 281
> 7300841
Length=1118
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 31/78 (39%), Positives = 43/78 (55%), Gaps = 2/78 (2%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CG L +VVAL GF +G+ G N + A + LGV+EI FI TFF T Y+S G +
Sbjct 201 CGTLTDVVALGAGFIDGLRLGENARLAAIHLGVKEIMRFIKTFFPSSKMSTFYESCGVTN 260
Query 61 VITTVF--GGNCRARRRL 76
+ + F N + R+L
Sbjct 261 AVASSFEIEANLHSGRKL 278
> SPAC23D3.04c
Length=373
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 30/67 (44%), Positives = 43/67 (64%), Gaps = 0/67 (0%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
GALKN+VA+A G +G+ G NTK+A++R+G+ E++ F FF +S G AD+
Sbjct 233 GALKNIVAVAAGIIDGLELGDNTKSAVMRIGLLEMQKFGRMFFDCKPLTMSEESCGIADL 292
Query 62 ITTVFGG 68
ITT GG
Sbjct 293 ITTCLGG 299
> 7291535
Length=349
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 0/71 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
C L+N++A A G +GM NTK I+R G E+ F+ F+ T ++S G +D
Sbjct 201 CSTLRNIIAFAAGCSDGMELNENTKGGIIRRGFLEMLQFVDVFYPGCRMGTFFESCGISD 260
Query 61 VITTVFGGNCR 71
++T+ + R
Sbjct 261 LVTSCYANRNR 271
> At2g40690
Length=433
Score = 44.7 bits (104), Expect = 5e-05, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
GALKNV+A+A G +GM G N+ AA++ G EI+ G T+ +G D+
Sbjct 272 GALKNVLAIAAGIVDGMNLGNNSMAALVSQGCSEIRWLATKVMG-AKPTTITGLSGTGDI 330
Query 62 ITTVFGGNCRAR 73
+ T F R R
Sbjct 331 MLTCFVNLSRNR 342
> 7302439
Length=580
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 21 GTNTKAAILRLGVEEIKTFIMTFFGD 46
G+N +LRL VE +K F+ FGD
Sbjct 391 GSNAILKLLRLPVERVKDFVPVTFGD 416
Lambda K H
0.326 0.142 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1162440328
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40