bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5621_orf2
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
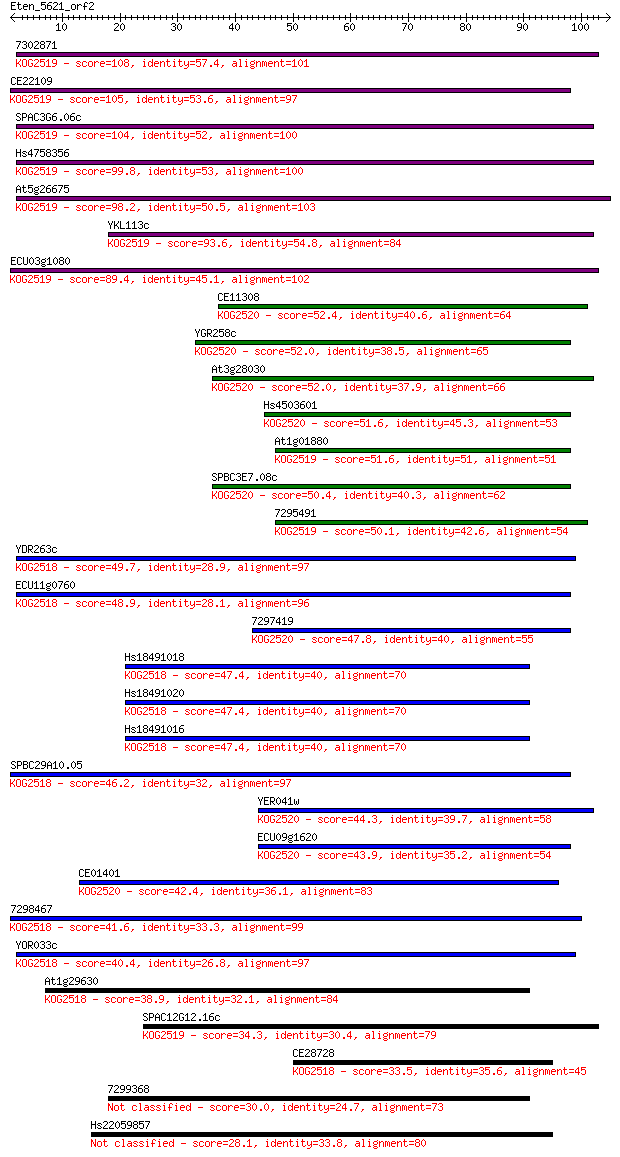
7302871 108 3e-24
CE22109 105 2e-23
SPAC3G6.06c 104 3e-23
Hs4758356 99.8 1e-21
At5g26675 98.2 3e-21
YKL113c 93.6 9e-20
ECU03g1080 89.4 1e-18
CE11308 52.4 2e-07
YGR258c 52.0 3e-07
At3g28030 52.0 3e-07
Hs4503601 51.6 3e-07
At1g01880 51.6 4e-07
SPBC3E7.08c 50.4 8e-07
7295491 50.1 1e-06
YDR263c 49.7 1e-06
ECU11g0760 48.9 2e-06
7297419 47.8 5e-06
Hs18491018 47.4 7e-06
Hs18491020 47.4 7e-06
Hs18491016 47.4 7e-06
SPBC29A10.05 46.2 2e-05
YER041w 44.3 5e-05
ECU09g1620 43.9 7e-05
CE01401 42.4 2e-04
7298467 41.6 4e-04
YOR033c 40.4 0.001
At1g29630 38.9 0.003
SPAC12G12.16c 34.3 0.063
CE28728 33.5 0.10
7299368 30.0 1.1
Hs22059857 28.1 4.2
> 7302871
Length=385
Score = 108 bits (270), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 58/101 (57%), Positives = 73/101 (72%), Gaps = 0/101 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R ERRE A A AA +AGD A + K R VRVT+E ++ K LL L+G+P + AP E
Sbjct 99 KRAERREEAEKALKAATDAGDDAGIEKFNRRLVRVTKEHAKEAKELLTLMGVPYVDAPCE 158
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDS 102
AEAQCAAL +AGKV+A ATED DALTFG+ LLR LT+S++
Sbjct 159 AEAQCAALVKAGKVYATATEDMDALTFGSTKLLRYLTYSEA 199
> CE22109
Length=382
Score = 105 bits (261), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 52/97 (53%), Positives = 68/97 (70%), Gaps = 0/97 (0%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG 60
E+R ERR A A A E GD E K R V+VT++QN++ KRLL L+G+P+++AP
Sbjct 98 EKRSERRAEAEKALTEAKEKGDVKEAEKFERRLVKVTKQQNDEAKRLLGLMGIPVVEAPC 157
Query 61 EAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
EAEAQCA L +AGKV+ TED DALTFG+ +LLR+
Sbjct 158 EAEAQCAHLVKAGKVFGTVTEDMDALTFGSTVLLRHF 194
> SPAC3G6.06c
Length=380
Score = 104 bits (260), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 52/100 (52%), Positives = 70/100 (70%), Gaps = 0/100 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R R + A E G A + + R+V+VTR+ N++ KRLL L+G+P + AP E
Sbjct 100 KRVARHQKAREDQEETKEVGTAEMVDRFAKRTVKVTRQHNDEAKRLLELMGIPFVNAPCE 159
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSD 101
AEAQCAALAR+GKV+AAA+ED D L F AP+LLR+LTFS+
Sbjct 160 AEAQCAALARSGKVYAAASEDMDTLCFQAPVLLRHLTFSE 199
> Hs4758356
Length=380
Score = 99.8 bits (247), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 53/100 (53%), Positives = 69/100 (69%), Gaps = 0/100 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R ERR A A AG E+ K R V+VT++ N++ K LL L+G+P L AP E
Sbjct 99 KRSERRAEAEKQLQQAQAAGAEQEVEKFTKRLVKVTKQHNDECKHLLSLMGIPYLDAPSE 158
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSD 101
AEA CAAL +AGKV+AAATED D LTFG+P+L+R+LT S+
Sbjct 159 AEASCAALVKAGKVYAAATEDMDCLTFGSPVLMRHLTASE 198
> At5g26675
Length=465
Score = 98.2 bits (243), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 52/115 (45%), Positives = 72/115 (62%), Gaps = 14/115 (12%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPL------ 55
+R +R A A A EAG+ ++ K R+V+VT++ N+D KRLL L+G+P+
Sbjct 100 KRYSKRADATADLTGAIEAGNKEDIEKYSKRTVKVTKQHNDDCKRLLRLMGVPVVEFEKK 159
Query 56 ------LQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDSSS 104
+QA EAEAQCAAL ++GKV+ A+ED D+LTFGAP LR+L D SS
Sbjct 160 TDVASYIQATSEAEAQCAALCKSGKVYGVASEDMDSLTFGAPKFLRHLM--DPSS 212
> YKL113c
Length=382
Score = 93.6 bits (231), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 46/84 (54%), Positives = 61/84 (72%), Gaps = 0/84 (0%)
Query 18 AEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWA 77
AEA E KQ R V+V++E NE+ ++LL L+G+P + AP EAEAQCA LA+ GKV+A
Sbjct 113 AEATTELEKMKQERRLVKVSKEHNEEAQKLLGLMGIPYIIAPTEAEAQCAELAKKGKVYA 172
Query 78 AATEDADALTFGAPILLRNLTFSD 101
AA+ED D L + P LLR+LTFS+
Sbjct 173 AASEDMDTLCYRTPFLLRHLTFSE 196
> ECU03g1080
Length=345
Score = 89.4 bits (220), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 46/102 (45%), Positives = 61/102 (59%), Gaps = 0/102 (0%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG 60
E+R+ERR AA A+E GD + R +VT ++ KRLL L+G+P AP
Sbjct 97 EKRKERRAAADREYREASEVGDKELMEMYDKRKTKVTGVHVDECKRLLGLMGIPFETAPS 156
Query 61 EAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDS 102
EAEA CA L + V+ ATED DALTFG+P++LRN + S
Sbjct 157 EAEAYCALLCKKKAVYGVATEDMDALTFGSPVVLRNFNGTQS 198
> CE11308
Length=829
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 38/64 (59%), Gaps = 1/64 (1%)
Query 37 TREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRN 96
T E D++ L G+P +++PGEAEAQC L R G V ++D+D FGA + R+
Sbjct 474 TPELYRDLQEFLTNAGIPWIESPGEAEAQCVELERLGLVDGVVSDDSDVWAFGAQHVYRH 533
Query 97 LTFS 100
+ FS
Sbjct 534 M-FS 536
> YGR258c
Length=1031
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 0/65 (0%)
Query 33 SVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPI 92
S VT + ++V+ LL G+P + AP EAEAQCA L + V T+D+D FG
Sbjct 764 SDEVTMDMIKEVQELLSRFGIPYITAPMEAEAQCAELLQLNLVDGIITDDSDVFLFGGTK 823
Query 93 LLRNL 97
+ +N+
Sbjct 824 IYKNM 828
> At3g28030
Length=1482
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 36 VTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLR 95
V+ E + + LL + G+P + AP EAEAQCA + ++ V T+D+D FGA + +
Sbjct 921 VSSEMFAECQELLQIFGIPYIIAPMEAEAQCAFMEQSNLVDGIVTDDSDVFLFGARSVYK 980
Query 96 NLTFSD 101
N+ F D
Sbjct 981 NI-FDD 985
> Hs4503601
Length=1186
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 45 KRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
+ LL L G+P +QAP EAEAQCA L + T+D+D FGA + RN
Sbjct 773 QELLRLFGIPYIQAPMEAEAQCAILDLTDQTSGTITDDSDIWLFGARHVYRNF 825
> At1g01880
Length=570
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/51 (50%), Positives = 35/51 (68%), Gaps = 0/51 (0%)
Query 47 LLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
LL LLG+P+L+A GEAEA CA L G V A T D+DA FGA +++++
Sbjct 126 LLELLGIPVLKANGEAEALCAQLNSQGFVDACITPDSDAFLFGAMCVIKDI 176
> SPBC3E7.08c
Length=1112
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 35/62 (56%), Gaps = 0/62 (0%)
Query 36 VTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLR 95
VT+ ++ + LL L GLP + AP EAEAQC+ L V T+D+D FG + R
Sbjct 752 VTQVMIKECQELLRLFGLPYIVAPQEAEAQCSKLLELKLVDGIVTDDSDVFLFGGTRVYR 811
Query 96 NL 97
N+
Sbjct 812 NM 813
> 7295491
Length=726
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 47 LLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFS 100
LLL +G+ +Q PGEAEA CA L + G V ++D+D +GA + RN + S
Sbjct 129 LLLSMGIQCVQGPGEAEAYCAFLNKHGLVDGVISQDSDCFAYGAVRVYRNFSVS 182
> YDR263c
Length=430
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 44/97 (45%), Gaps = 0/97 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
RRR++R A AG+ + +SV +T E + + L +P + AP E
Sbjct 91 RRRKKRLENEMIAKKLWSAGNRYNAMEYFQKSVDITPEMAKCIIDYCKLHSIPYIVAPFE 150
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT 98
A+ Q L + G + +ED+D L FG L+ L
Sbjct 151 ADPQMVYLEKMGLIQGIISEDSDLLVFGCKTLITKLN 187
> ECU11g0760
Length=366
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 49/96 (51%), Gaps = 0/96 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+RR R+E + A D A+ + + + + VTRE D+ R+L + + + +P E
Sbjct 91 KRRIRKEKSRKEAEYWLMRNDPAKAKGFMRQCIAVTREVVSDIARMLERINVEYIISPYE 150
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
++AQ L R G + TED+D + +G+ +L
Sbjct 151 SDAQLCFLQRIGYIDCILTEDSDLIPYGSSKVLYKF 186
> 7297419
Length=1236
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 43 DVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
D + LL L G+P + AP EAEAQCA L T+D+D FG + +N
Sbjct 862 DCQELLRLFGIPYIVAPMEAEAQCAFLNATDLTHGTITDDSDIWLFGGRTVYKNF 916
> Hs18491018
Length=803
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 21 GDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAAT 80
G +E R+ RS+ +T V + G+ L AP EA+AQ A L +AG V A T
Sbjct 110 GKVSEARECFTRSINITHAMAHKVIKAARSQGVDCLVAPYEADAQLAYLNKAGIVQAIIT 169
Query 81 EDADALTFGA 90
ED+D L FG
Sbjct 170 EDSDLLAFGC 179
> Hs18491020
Length=846
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 21 GDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAAT 80
G +E R+ RS+ +T V + G+ L AP EA+AQ A L +AG V A T
Sbjct 110 GKVSEARECFTRSINITHAMAHKVIKAARSQGVDCLVAPYEADAQLAYLNKAGIVQAIIT 169
Query 81 EDADALTFGA 90
ED+D L FG
Sbjct 170 EDSDLLAFGC 179
> Hs18491016
Length=846
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 21 GDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAAT 80
G +E R+ RS+ +T V + G+ L AP EA+AQ A L +AG V A T
Sbjct 110 GKVSEARECFTRSINITHAMAHKVIKAARSQGVDCLVAPYEADAQLAYLNKAGIVQAIIT 169
Query 81 EDADALTFGA 90
ED+D L FG
Sbjct 170 EDSDLLAFGC 179
> SPBC29A10.05
Length=571
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 50/100 (50%), Gaps = 6/100 (6%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLL---GLPLLQ 57
++R+ERR+ A + G ++ Q +R V VT E +L++ L G+ +
Sbjct 90 QKRKERRQEAFELGKKLWDEGKKSQAIMQFSRCVDVTPEM---AWKLIIALREHGIESIV 146
Query 58 APGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
AP EA+AQ L + + TED+D L FGA +L +
Sbjct 147 APYEADAQLVYLEKENIIDGIITEDSDMLVFGAQTVLFKM 186
> YER041w
Length=759
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 44 VKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT-FSD 101
V++LL L+ + + A GE EAQC L +G V + D+D L FG +L+N + F D
Sbjct 176 VRKLLDLMNISYVIACGEGEAQCVWLQVSGAVDFILSNDSDTLVFGGEKILKNYSKFYD 234
> ECU09g1620
Length=562
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 1/54 (1%)
Query 44 VKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
VK +L + +P + AP EA+ QC + V TED D L +G + RN
Sbjct 392 VKSVLEVFNIPYVDAPMEADGQCGFMCHNNVVDGVITEDNDVLLYGGTV-YRNF 444
> CE01401
Length=434
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 42/88 (47%), Gaps = 5/88 (5%)
Query 13 AAAAAAEAGDAAELRKQLARSVRVTREQN--EDVKR---LLLLLGLPLLQAPGEAEAQCA 67
A+++A E+ D E + RS + N + V + LL LG+ ++ APG+ EAQCA
Sbjct 80 ASSSAHESKDQNEFVPRKRRSFGDSPFTNLVDHVYKTNALLTELGIKVIIAPGDGEAQCA 139
Query 68 ALARAGKVWAAATEDADALTFGAPILLR 95
L G T D D FG L R
Sbjct 140 RLEDLGVTSGCITTDFDYFLFGGKNLYR 167
> 7298467
Length=732
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 33/99 (33%), Positives = 46/99 (46%), Gaps = 4/99 (4%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG 60
+RRR+ R+ + AA G E R + R V VT + + R + + AP
Sbjct 90 KRRRDSRKQSKERAAELLRLGRIEEARSHMRRCVDVTHDMALRLIRECRSRNVDCIVAPY 149
Query 61 EAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTF 99
EA+AQ A L RA TED+D FGA +N+ F
Sbjct 150 EADAQMAWLNRADVAQYIITEDSDLTLFGA----KNIIF 184
> YOR033c
Length=702
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 43/97 (44%), Gaps = 0/97 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+RR++R+ A A G+ + V +T E + + L G+ + AP E
Sbjct 91 KRRDKRKENKAIAERLWACGEKKNAMDYFQKCVDITPEMAKCIICYCKLNGIRYIVAPFE 150
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT 98
A++Q L + V +ED+D L FG L+ L
Sbjct 151 ADSQMVYLEQKNIVQGIISEDSDLLVFGCRRLITKLN 187
> At1g29630
Length=317
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 41/85 (48%), Gaps = 12/85 (14%)
Query 7 REAAAAAAAAAAEAGDAAELRKQLARS-VRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQ 65
EA ++AA A ++ +A ++V R++N D + AP EA+AQ
Sbjct 106 HEANGNSSAAYECYSKAVDISPSIAHELIQVLRQENVDY-----------VVAPYEADAQ 154
Query 66 CAALARAGKVWAAATEDADALTFGA 90
A LA +V A TED+D + FG
Sbjct 155 MAFLAITKQVDAIITEDSDLIPFGC 179
> SPAC12G12.16c
Length=496
Score = 34.3 bits (77), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 24 AELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAP--GEAEAQCAALARAGKVWAAATE 81
EL K R R + + ++ +L +LG+P +P EAEA +A+++ +A AT+
Sbjct 261 TELDKLERRLYRPSPQNIFEIFEILKILGIPASFSPIGVEAEAFASAISQNNLAYAVATQ 320
Query 82 DADALTFGAPILLRNLTFSDS 102
D D L G+ ++ L +D+
Sbjct 321 DTDVLLLGSSMISNFLDLNDN 341
> CE28728
Length=639
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 50 LLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILL 94
+ + ++ AP EA+AQ A L + V A TED+D + FG ++
Sbjct 140 MTNVDIVVAPYEADAQLAYLVQEKLVDAVITEDSDLIVFGCEMIY 184
> 7299368
Length=313
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 35/73 (47%), Gaps = 0/73 (0%)
Query 18 AEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWA 77
A GD +++ ++ RS ++ NED ++ +L + + +AQ +AL R +
Sbjct 135 ASEGDVSKMFRKAVRSTFSSQSTNEDQSKIDILDLVASTSSSSTGDAQKSALNRKTPTFE 194
Query 78 AATEDADALTFGA 90
+ D +TFG
Sbjct 195 SHFIDVGTITFGG 207
> Hs22059857
Length=334
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 36/97 (37%), Gaps = 21/97 (21%)
Query 15 AAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG-------------- 60
AAA G R Q +V R+ + V+ + LG+P+ APG
Sbjct 79 GAAAVPGSDLTHRTQAGHKTQVPRKAHSSVRLTMQDLGIPVEGAPGRDGTAPSAAQTPGA 138
Query 61 -EAEAQCAALARAG--KVWAAATEDADALTFGAPILL 94
A A L G + W A+ED GAP L
Sbjct 139 SSATTHQATLPTQGICQTWKPASEDPK----GAPQCL 171
Lambda K H
0.315 0.125 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40