bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5677_orf1
Length=155
Score E
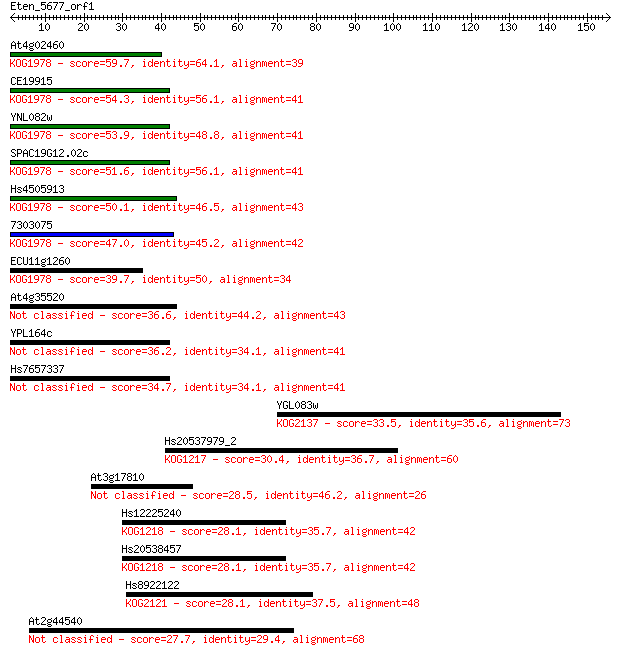
Sequences producing significant alignments: (Bits) Value
At4g02460 59.7 2e-09
CE19915 54.3 9e-08
YNL082w 53.9 1e-07
SPAC19G12.02c 51.6 5e-07
Hs4505913 50.1 2e-06
7303075 47.0 2e-05
ECU11g1260 39.7 0.002
At4g35520 36.6 0.018
YPL164c 36.2 0.029
Hs7657337 34.7 0.066
YGL083w 33.5 0.17
Hs20537979_2 30.4 1.3
At3g17810 28.5 6.0
Hs12225240 28.1 7.2
Hs20538457 28.1 7.3
Hs8922122 28.1 7.6
At2g44540 27.7 9.3
> At4g02460
Length=779
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 25/39 (64%), Positives = 30/39 (76%), Gaps = 0/39 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLV 39
VMIGDPL + I+ +ADL+ P+NCPHGRPTMRHLV
Sbjct 714 VMIGDPLRKNEMQKIVEHLADLESPWNCPHGRPTMRHLV 752
> CE19915
Length=805
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 23/41 (56%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLVDL 41
VMIG PL I+R +A L P+NCPHGRPT+RHL L
Sbjct 758 VMIGKPLNQREMTQIIRHLAKLDQPWNCPHGRPTIRHLASL 798
> YNL082w
Length=904
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 20/41 (48%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLVDL 41
+MIG PL ++ +++L P+NCPHGRPTMRHL++L
Sbjct 852 IMIGKPLNKKTMTRVVHNLSELDKPWNCPHGRPTMRHLMEL 892
> SPAC19G12.02c
Length=794
Score = 51.6 bits (122), Expect = 5e-07, Method: Composition-based stats.
Identities = 23/41 (56%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLVDL 41
VMIG L I+R +A+L P+NCPHGRPTMRHL+ L
Sbjct 751 VMIGRALTISEMNTIVRHLAELSKPWNCPHGRPTMRHLLRL 791
> Hs4505913
Length=862
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLVDLGA 43
VMIG L ++ M ++ P+NCPHGRPTMRH+ +LG
Sbjct 816 VMIGTALNTSEMKKLITHMGEMDHPWNCPHGRPTMRHIANLGV 858
> 7303075
Length=899
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 19/43 (44%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 1 VMIGDPLPPHRQLA-ILRRMADLQLPFNCPHGRPTMRHLVDLG 42
VMIG L + + ++ +M +++ P+NCPHGRPTMRHL+++
Sbjct 842 VMIGTALSRNTTMRRLITQMGEIEQPWNCPHGRPTMRHLINIA 884
> ECU11g1260
Length=630
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 17/34 (50%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPT 34
VMIGD L I++ +A L+ P+ CPHGRPT
Sbjct 588 VMIGDVLSMADMKRIVKSLASLERPWKCPHGRPT 621
> At4g35520
Length=1151
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 19/43 (44%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLVDLGA 43
+M GD L P I+ + L F C HGRPT LVDL A
Sbjct 1073 IMFGDSLLPSECSLIIDGLKQTSLCFQCAHGRPTTVPLVDLKA 1115
> YPL164c
Length=715
Score = 36.2 bits (82), Expect = 0.029, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLVDL 41
VM GD L + ++ +++ PF C HGRP+M + +L
Sbjct 674 VMFGDELTRQECIILISKLSRCHNPFECAHGRPSMVPIAEL 714
> Hs7657337
Length=1429
Score = 34.7 bits (78), Expect = 0.066, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 1 VMIGDPLPPHRQLAILRRMADLQLPFNCPHGRPTMRHLVDL 41
+ D L ++ ++ QLPF C HGRP+M L D+
Sbjct 1342 IKFNDGLSLQESCRLIEALSSCQLPFQCAHGRPSMLPLADI 1382
> YGL083w
Length=804
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 38/87 (43%), Gaps = 16/87 (18%)
Query 70 PLDPPSCLRGSKNATFAPSDDSGNCQE----------GSTGAARQKRRPSSDWKGMDSRP 119
P PS GSK T S+D+ + + STG+ RQ PS W M+S P
Sbjct 705 PPQTPSSRTGSKVMTKGGSNDASSTKVEEEFNEFQSFSSTGSIRQTSAPSDVW--MNSTP 762
Query 120 TKIPTCVA----PRGGIVTGRNEQRGD 142
+ PT + P G ++ + +R D
Sbjct 763 SPTPTSASSTNLPPGFSISLQPNKRKD 789
> Hs20537979_2
Length=1158
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 41 LGASAGQANERLSNGSSRAGIQVVQPNGVPLDPPSCLRGSKNATFAPSDDSGNCQEGSTG 100
LGA A A +R + S+ + I+V QP+GV +PP CL + + P G+CQ+ G
Sbjct 461 LGAVALYACDRGYSLSAPSRIRVCQPHGVWSEPPQCLEIDECRS-QPCLHGGSCQDRVAG 519
> At3g17810
Length=426
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 17/26 (65%), Gaps = 4/26 (15%)
Query 22 LQLPFNCPHGRPTMRHLVDLGASAGQ 47
L++ F+CPHG P R +GA+ GQ
Sbjct 185 LEINFSCPHGMPERR----MGAAVGQ 206
> Hs12225240
Length=2570
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 30 HGRPTMRHLVDLGASAGQANERLSNGSSRAGIQVVQPNGVPL 71
HG+ T+ L+ G AN+ L+ S G ++ P GVPL
Sbjct 581 HGQLTVEKLISKGRILTMANQVLAVNISEEGRILLGPEGVPL 622
> Hs20538457
Length=2570
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 30 HGRPTMRHLVDLGASAGQANERLSNGSSRAGIQVVQPNGVPL 71
HG+ T+ L+ G AN+ L+ S G ++ P GVPL
Sbjct 581 HGQLTVEKLISKGRILTMANQVLAVNISEEGRILLGPEGVPL 622
> Hs8922122
Length=363
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Query 31 GRPTMRHLVDLGASAGQANERLSNGSSRAGIQVVQPNGVPLDPPSCLR 78
G+ + L DLG G A +L N GI VV NGV + P L+
Sbjct 198 GKLNAQKLKDLGVPPGPAYGKLKN-----GISVVLENGVTISPQDVLK 240
> At2g44540
Length=491
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 28/74 (37%), Gaps = 6/74 (8%)
Query 6 PLPPHRQLAILRRMADLQLPFNCPHG------RPTMRHLVDLGASAGQANERLSNGSSRA 59
P PH + A + + + P C G P V +GA G NE G R+
Sbjct 409 PKKPHHRAASIVSIRKDKTPVTCSGGYDKWYNNPAPNPNVLMGALVGGPNENDVYGDERS 468
Query 60 GIQVVQPNGVPLDP 73
Q +P V + P
Sbjct 469 NFQQAEPATVTVAP 482
Lambda K H
0.318 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40