bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5699_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
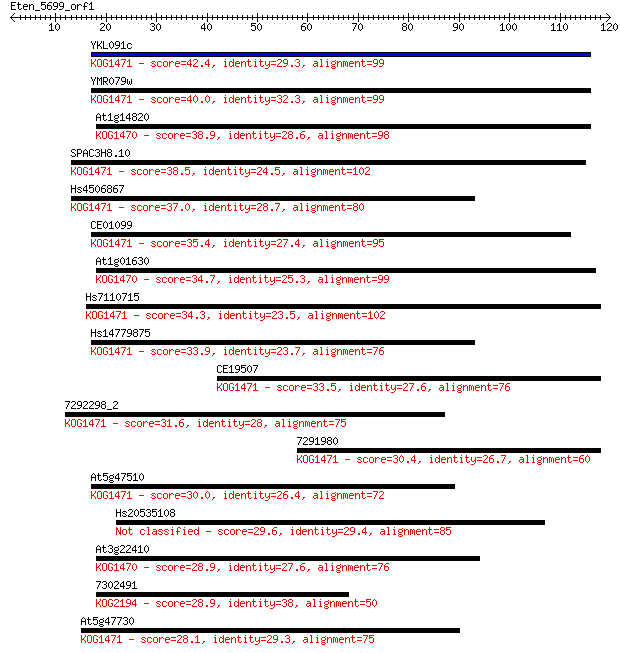
YKL091c 42.4 2e-04
YMR079w 40.0 0.001
At1g14820 38.9 0.002
SPAC3H8.10 38.5 0.003
Hs4506867 37.0 0.008
CE01099 35.4 0.028
At1g01630 34.7 0.039
Hs7110715 34.3 0.051
Hs14779875 33.9 0.068
CE19507 33.5 0.096
7292298_2 31.6 0.38
7291980 30.4 0.86
At5g47510 30.0 1.0
Hs20535108 29.6 1.3
At3g22410 28.9 2.4
7302491 28.9 2.4
At5g47730 28.1 3.7
> YKL091c
Length=310
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 51/101 (50%), Gaps = 4/101 (3%)
Query 17 VESWCILCDLSGISISS--FPLPLLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKPLV 74
+E+ C + DL GIS+S+ L + + + Q Y R+ +FYI+H+P F + K +
Sbjct 172 IETSCTVLDLKGISLSNAYHVLSYIKDVADISQNYYPERMGKFYIIHSPFGFSTMFKMVK 231
Query 75 ASLPSTTAKKLRVFTSFDEWQQERRAQFTSPQIEEKFGGTA 115
L T K+ + S +++E Q + K+GGT+
Sbjct 232 PFLDPVTVSKIFILGS--SYKKELLKQIPIENLPVKYGGTS 270
> YMR079w
Length=304
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/108 (29%), Positives = 50/108 (46%), Gaps = 18/108 (16%)
Query 17 VESWCILCDLSGISISSF--PLPLLFKLMQLIQGPYRGRLYRFYILHAP-------RLFH 67
VE+ C + DL GISISS + + + + Q Y R+ +FYI++AP RLF
Sbjct 170 VETSCTIMDLKGISISSAYSVMSYVREASYISQNYYPERMGKFYIINAPFGFSTAFRLFK 229
Query 68 FVAKPLVASLPSTTAKKLRVFTSFDEWQQERRAQFTSPQIEEKFGGTA 115
P+ S ++F +Q+E Q + + KFGG +
Sbjct 230 PFLDPVTVS---------KIFILGSSYQKELLKQIPAENLPVKFGGKS 268
> At1g14820
Length=252
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 46/98 (46%), Gaps = 1/98 (1%)
Query 18 ESWCILCDLSGISISSFPLPLLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKPLVASL 77
E + DL+ I+ + L Q +Q Y RL + YILH P F V K + L
Sbjct 133 EKLVAVIDLANITYKNLDARGLITGFQFLQSYYPERLAKCYILHMPGFFVTVWKFVCRFL 192
Query 78 PSTTAKKLRVFTSFDEWQQERRAQFTSPQIEEKFGGTA 115
T +K+ + T +E Q++ + + + E++GG A
Sbjct 193 EKATQEKIVIVTDGEE-QRKFEEEIGADALPEEYGGRA 229
> SPAC3H8.10
Length=286
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/104 (24%), Positives = 45/104 (43%), Gaps = 4/104 (3%)
Query 13 VAGRVESWCILCDLSGISISSFP--LPLLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVA 70
G +E+ C + DL G+ I+S + + + Q Y R+ +FY+++AP F
Sbjct 161 AGGLIETSCTIMDLKGVGITSIHSVYSYIRQASSISQDYYPERMGKFYVINAPWGFSSAF 220
Query 71 KPLVASLPSTTAKKLRVFTSFDEWQQERRAQFTSPQIEEKFGGT 114
+ L T KK+ + S ++ Q + + K GG
Sbjct 221 NLIKGFLDEATVKKIHILGS--NYKSALLEQIPADNLPAKLGGN 262
> Hs4506867
Length=715
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 41/84 (48%), Gaps = 4/84 (4%)
Query 13 VAGR-VESWCILCDLSGISISSFPLP---LLFKLMQLIQGPYRGRLYRFYILHAPRLFHF 68
V GR + SW L DL G+++ P L +++++++ Y L R IL APR+F
Sbjct 384 VFGRPISSWTCLVDLEGLNMRHLWRPGVKALLRIIEVVEANYPETLGRLLILRAPRVFPV 443
Query 69 VAKPLVASLPSTTAKKLRVFTSFD 92
+ + + T +K ++ D
Sbjct 444 LWTLVSPFIDDNTRRKFLIYAGND 467
> CE01099
Length=743
Score = 35.4 bits (80), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 48/99 (48%), Gaps = 9/99 (9%)
Query 17 VESWCILCDLSGISISSFPLP---LLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKPL 73
+ SW ++ DL G+S+ P L K++++++ Y + + ++ APR+F + +
Sbjct 413 ISSWSLVVDLDGLSMRHLWRPGVQCLLKIIEIVEANYPETMGQVLVVRAPRVFPVLWTLI 472
Query 74 VASLPSTTAKKLRVF-TSFDEWQQERRAQFTSPQIEEKF 111
+ T KK V S + ++E R IEEKF
Sbjct 473 SPFIDEKTRKKFMVSGGSGGDLKEELRK-----HIEEKF 506
> At1g01630
Length=255
Score = 34.7 bits (78), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 42/99 (42%), Gaps = 1/99 (1%)
Query 18 ESWCILCDLSGISISSFPLPLLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKPLVASL 77
E + + DL G S+ + + +Q Y RL + YI+HAP +F K + +
Sbjct 151 EKFVAIGDLQGWGYSNCDIRGYLAALSTLQDCYPERLGKLYIVHAPYIFMTAWKVIYPFI 210
Query 78 PSTTAKKLRVFTSFDEWQQERRAQFTSPQIEEKFGGTAP 116
+ T KK+ VF + Q+ + +GG P
Sbjct 211 DANTKKKI-VFVENKKLTPTLLEDIDESQLPDIYGGKLP 248
> Hs7110715
Length=403
Score = 34.3 bits (77), Expect = 0.051, Method: Composition-based stats.
Identities = 24/105 (22%), Positives = 47/105 (44%), Gaps = 5/105 (4%)
Query 16 RVESWCILCDLSGISISSFPLPLL---FKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKP 72
+VE+ I+ D G+ + P + + + + + Y L R +++ AP+LF
Sbjct 145 KVETITIIYDCEGLGLKHLWKPAVEAYGEFLCMFEENYPETLKRLFVVKAPKLFPVAYNL 204
Query 73 LVASLPSTTAKKLRVFTSFDEWQQERRAQFTSPQIEEKFGGTAPD 117
+ L T KK+ V + W++ + Q+ ++GGT D
Sbjct 205 IKPFLSEDTRKKIMVLGA--NWKEVLLKHISPDQVPVEYGGTMTD 247
> Hs14779875
Length=696
Score = 33.9 bits (76), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 18/79 (22%), Positives = 39/79 (49%), Gaps = 3/79 (3%)
Query 17 VESWCILCDLSGISISSFPLP---LLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKPL 73
+ SW L DL G+++ P L +++++++ Y L R I+ APR+F + +
Sbjct 376 ISSWTCLLDLEGLNMRHLWRPGVKALLRMIEVVEDNYPETLGRLLIVRAPRVFPVLWTLI 435
Query 74 VASLPSTTAKKLRVFTSFD 92
+ T +K +++ +
Sbjct 436 SPFINENTRRKFLIYSGSN 454
> CE19507
Length=396
Score = 33.5 bits (75), Expect = 0.096, Method: Composition-based stats.
Identities = 21/87 (24%), Positives = 38/87 (43%), Gaps = 11/87 (12%)
Query 42 LMQLIQGPYR---GRLYRFY--------ILHAPRLFHFVAKPLVASLPSTTAKKLRVFTS 90
L+ ++ GPYR +Y Y +++AP + K + LP T K+R+ +
Sbjct 157 LISIVTGPYRILWASVYTAYPEWINTLFLINAPSFMTLLWKAIGPLLPERTRNKVRICSG 216
Query 91 FDEWQQERRAQFTSPQIEEKFGGTAPD 117
+W+ + I + +GGT D
Sbjct 217 NSDWKTSVQKHAHIDNIPKHWGGTLVD 243
> 7292298_2
Length=212
Score = 31.6 bits (70), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 0/75 (0%)
Query 12 CVAGRVESWCILCDLSGISISSFPLPLLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAK 71
C + CI+ DL+ S S L+ L+ L+ + RL I+++P LF +
Sbjct 117 CFEEVTDRLCIVFDLAEFSTSCMDYQLVQNLIWLLGKHFPERLGVCLIINSPGLFSTIWP 176
Query 72 PLVASLPSTTAKKLR 86
+ L TAKK++
Sbjct 177 AIRVLLDDNTAKKVK 191
> 7291980
Length=385
Score = 30.4 bits (67), Expect = 0.86, Method: Composition-based stats.
Identities = 16/61 (26%), Positives = 28/61 (45%), Gaps = 1/61 (1%)
Query 58 YILHAPRLFHFVAKPLVASLPSTTAKKLRVFTS-FDEWQQERRAQFTSPQIEEKFGGTAP 116
YI++AP+LF + L T K+ ++ S D WQ++ + + +GG
Sbjct 167 YIINAPKLFSVAFNIVKKFLDENTTSKIVIYKSGVDRWQEQLFSHVNRKAFPKAWGGEMV 226
Query 117 D 117
D
Sbjct 227 D 227
> At5g47510
Length=403
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 17 VESWCILCDLSGISISSFPLP---LLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKPL 73
V S + D+SG+ +S+F P L ++ ++ Y L+R ++++A F + L
Sbjct 163 VSSTTTILDVSGVGMSNFSKPARSLFMEIQKIDSNYYPETLHRLFVVNASSGFRMLWLAL 222
Query 74 VASLPSTTAKKLRVF 88
L + T K++V
Sbjct 223 KTFLDARTLAKVQVL 237
> Hs20535108
Length=193
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 25/101 (24%), Positives = 41/101 (40%), Gaps = 16/101 (15%)
Query 22 ILCDLSGISISSFPLPLLFKLMQLIQGPYRG------------RLYRFYILHAPRLFHFV 69
IL +GI ++S P+ +FK+ G L+ F +L AP+ +
Sbjct 89 ILSSYAGILMNSIPIEEVFKIYGADSSADSGTIKVPRVSSLCLSLHPFAMLTAPKAAAYA 148
Query 70 AKPLVASLPSTTAKKLRVFTSFD----EWQQERRAQFTSPQ 106
K V S TT K ++ + EW+ +R T P+
Sbjct 149 RKQSVKSRKVTTNKNATSISAKEANATEWKSSQRFSDTQPK 189
> At3g22410
Length=400
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 37/76 (48%), Gaps = 0/76 (0%)
Query 18 ESWCILCDLSGISISSFPLPLLFKLMQLIQGPYRGRLYRFYILHAPRLFHFVAKPLVASL 77
+S+ +L D S SS LL +++I Y RLY+ +I+ P F ++ K + +
Sbjct 132 QSFVLLFDASFFRSSSAFANLLLATLKIIADNYPCRLYKAFIIDPPSFFSYLWKGVRPFV 191
Query 78 PSTTAKKLRVFTSFDE 93
+TA + +DE
Sbjct 192 ELSTATMILSSLDYDE 207
> 7302491
Length=891
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 26/52 (50%), Gaps = 7/52 (13%)
Query 18 ESWCILCDLSGISISSFPLPLLFKLMQLIQGPYR--GRLYRFYILHAPRLFH 67
+S I+C L GI++ F + M I PYR + RF +LH R FH
Sbjct 634 KSKTIICGLMGITLLCFIIA-----MTPIGFPYRPETNVQRFAVLHTKRTFH 680
> At5g47730
Length=341
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 40/80 (50%), Gaps = 8/80 (10%)
Query 15 GRVESWCI-LCDLSGISISSFPLPLLFKLMQLIQG-PYRGRLYRFYILHAPRLFHF---V 69
GR + C+ + D++G+ +S+ L ++ I Y + +Y+++AP +F V
Sbjct 146 GRPITTCVKVLDMTGLKLSALSQIKLVTIISTIDDLNYPEKTNTYYVVNAPYIFSACWKV 205
Query 70 AKPLVASLPSTTAKKLRVFT 89
KPL L T KK+ V +
Sbjct 206 VKPL---LQERTRKKVHVLS 222
Lambda K H
0.331 0.144 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40