bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
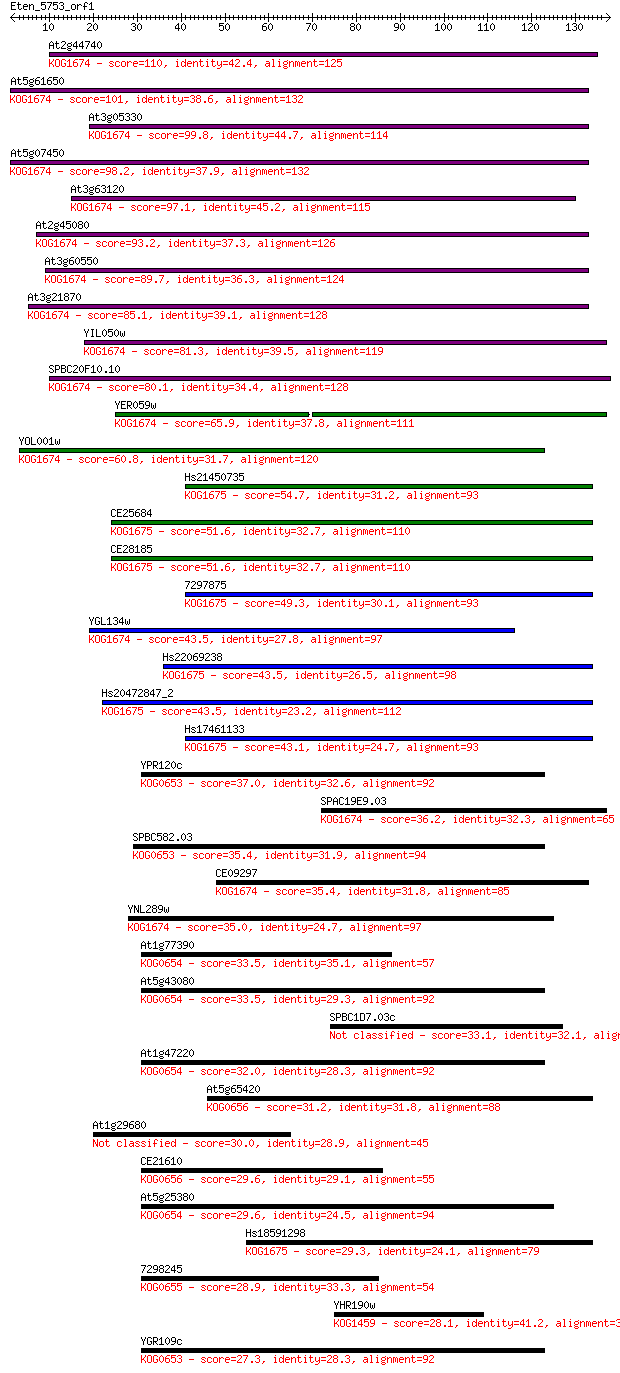
Query= Eten_5753_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
At2g44740 110 8e-25
At5g61650 101 3e-22
At3g05330 99.8 1e-21
At5g07450 98.2 4e-21
At3g63120 97.1 1e-20
At2g45080 93.2 1e-19
At3g60550 89.7 2e-18
At3g21870 85.1 3e-17
YIL050w 81.3 5e-16
SPBC20F10.10 80.1 1e-15
YER059w 65.9 2e-11
YOL001w 60.8 8e-10
Hs21450735 54.7 5e-08
CE25684 51.6 5e-07
CE28185 51.6 5e-07
7297875 49.3 2e-06
YGL134w 43.5 1e-04
Hs22069238 43.5 1e-04
Hs20472847_2 43.5 1e-04
Hs17461133 43.1 1e-04
YPR120c 37.0 0.010
SPAC19E9.03 36.2 0.020
SPBC582.03 35.4 0.031
CE09297 35.4 0.032
YNL289w 35.0 0.040
At1g77390 33.5 0.12
At5g43080 33.5 0.12
SPBC1D7.03c 33.1 0.15
At1g47220 32.0 0.31
At5g65420 31.2 0.63
At1g29680 30.0 1.2
CE21610 29.6 1.7
At5g25380 29.6 1.8
Hs18591298 29.3 2.0
7298245 28.9 2.7
YHR190w 28.1 4.7
YGR109c 27.3 9.7
> At2g44740
Length=202
Score = 110 bits (275), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 53/125 (42%), Positives = 76/125 (60%), Gaps = 0/125 (0%)
Query 10 SSEGCGAPCFLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPL 69
+++ F + P I++ YLER+ ++ CS CFV+A VY+DR + +
Sbjct 35 ATQSQRVSVFHGLSRPTITIQSYLERIFKYANCSPSCFVVAYVYLDRFTHRQPSLPINSF 94
Query 70 NLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMPCEF 129
N+HRL +T++MVA KF DD +Y+NAYYAKVGG+ +EMN LE L L F L+V P F
Sbjct 95 NVHRLLITSVMVAAKFLDDLYYNNAYYAKVGGISTKEMNFLELDFLFGLGFELNVTPNTF 154
Query 130 DKYFK 134
+ YF
Sbjct 155 NAYFS 159
> At5g61650
Length=219
Score = 101 bits (252), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 51/134 (38%), Positives = 78/134 (58%), Gaps = 2/134 (1%)
Query 1 LLQHLGVQNS--SEGCGAPCFLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPL 58
LLQ + N S+ F T+P IS+ YLER+ + CS C+++A +Y+DR +
Sbjct 32 LLQRVSETNDNLSQKQKPSSFTGVTKPSISIRSYLERIFEYANCSYSCYIVAYIYLDRFV 91
Query 59 QMNNHVWLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRML 118
+ + + N+HRL +T+++V+ KF DD Y+N YYAKVGG+ +EMN LE L +
Sbjct 92 KKQPFLPINSFNVHRLIITSVLVSAKFMDDLSYNNEYYAKVGGISREEMNMLELDFLFGI 151
Query 119 HFRLHVMPCEFDKY 132
F L+V F+ Y
Sbjct 152 GFELNVTVSTFNNY 165
> At3g05330
Length=588
Score = 99.8 bits (247), Expect = 1e-21, Method: Composition-based stats.
Identities = 51/118 (43%), Positives = 71/118 (60%), Gaps = 4/118 (3%)
Query 19 FLSATEPMISMPDYLERLARFFQCSGECFVLALVYI----DRPLQMNNHVWLCPLNLHRL 74
F + P +S+ Y ER+ R+ QCS CFV A YI RP + L LN+HRL
Sbjct 441 FHGSKAPSLSIYRYTERIHRYAQCSPVCFVAAFAYILRYLQRPEATSTARRLTSLNVHRL 500
Query 75 AVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMPCEFDKY 132
+T+L+VA KF + Y+NAYYAK+GG+ +EMN LE T L + FRL++ F+K+
Sbjct 501 LITSLLVAAKFLERQCYNNAYYAKIGGVSTEEMNRLERTFLVDVDFRLYITTETFEKH 558
> At5g07450
Length=216
Score = 98.2 bits (243), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 50/138 (36%), Positives = 81/138 (58%), Gaps = 6/138 (4%)
Query 1 LLQHLGVQNSS------EGCGAPCFLSATEPMISMPDYLERLARFFQCSGECFVLALVYI 54
LLQ + N E F + T+P IS+ Y+ER+ ++ CS C+++A +Y+
Sbjct 30 LLQRVSETNDDLSRPFREHKRISAFNAVTKPSISIRSYMERIFKYADCSDSCYIVAYIYL 89
Query 55 DRPLQMNNHVWLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATL 114
DR +Q + + N+HRL +T+++V+ KF DD Y+NA+YAKVGG+ +EMN LE
Sbjct 90 DRFIQKQPLLPIDSSNVHRLIITSVLVSAKFMDDLCYNNAFYAKVGGITTEEMNLLELDF 149
Query 115 LRMLHFRLHVMPCEFDKY 132
L + F+L+V ++ Y
Sbjct 150 LFGIGFQLNVTISTYNDY 167
> At3g63120
Length=221
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 52/119 (43%), Positives = 73/119 (61%), Gaps = 5/119 (4%)
Query 15 GAP----CFLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLN 70
G+P F + P IS+ YL+R+ ++ CS CFV+A +YID L L PLN
Sbjct 57 GSPDSVTVFDGRSPPEISIAHYLDRIFKYSCCSPSCFVIAHIYIDHFLH-KTRALLKPLN 115
Query 71 LHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMPCEF 129
+HRL +T +M+A K DD +++NAYYA+VGG+ +E+N LE LL L F+L V P F
Sbjct 116 VHRLIITTVMLAAKVFDDRYFNNAYYARVGGVTTRELNRLEMELLFTLDFKLQVDPQTF 174
> At2g45080
Length=222
Score = 93.2 bits (230), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 47/127 (37%), Positives = 73/127 (57%), Gaps = 1/127 (0%)
Query 7 VQNSSEGCGAPCFLSATE-PMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVW 65
+ S G G E P +++ YLER+ R+ + +V+A VYIDR Q N
Sbjct 48 ISRSYGGFGKTRVFDCREIPDMTIQSYLERIFRYTKAGPSVYVVAYVYIDRFCQNNQGFR 107
Query 66 LCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVM 125
+ N+HRL +T +M+A K+ +D Y N+Y+AKVGGL +++N+LE L ++ F+LHV
Sbjct 108 ISLTNVHRLLITTIMIASKYVEDMNYKNSYFAKVGGLETEDLNNLELEFLFLMGFKLHVN 167
Query 126 PCEFDKY 132
F+ Y
Sbjct 168 VSVFESY 174
> At3g60550
Length=230
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 45/124 (36%), Positives = 70/124 (56%), Gaps = 0/124 (0%)
Query 9 NSSEGCGAPCFLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCP 68
+S G F P +++ YL R+ R+ + +V+A VYIDR Q N +
Sbjct 57 SSGAGGKTQIFDCREIPDMTIQSYLGRIFRYTKAGPSVYVVAYVYIDRFCQTNPGFRISL 116
Query 69 LNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMPCE 128
N+HRL +T +M+A K+ +D Y N+Y+AKVGGL +++N LE L ++ F+LHV
Sbjct 117 TNVHRLLITTIMIASKYVEDLNYRNSYFAKVGGLETEDLNKLELEFLFLMGFKLHVNVSV 176
Query 129 FDKY 132
F+ Y
Sbjct 177 FESY 180
> At3g21870
Length=210
Score = 85.1 bits (209), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 50/128 (39%), Positives = 71/128 (55%), Gaps = 0/128 (0%)
Query 5 LGVQNSSEGCGAPCFLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHV 64
L Q G F P IS+ YLER+ ++ +CS CFV+ VYIDR +
Sbjct 45 LAKQTKGFGKSLEAFHGVRAPSISIAKYLERIYKYTKCSPACFVVGYVYIDRLAHKHPGS 104
Query 65 WLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHV 124
+ LN+HRL VT +M+A K DD Y+N +YA+VGG+ ++N +E LL +L FR+ V
Sbjct 105 LVVSLNVHRLLVTCVMIAAKILDDVHYNNEFYARVGGVSNADLNKMELELLFLLDFRVTV 164
Query 125 MPCEFDKY 132
F+ Y
Sbjct 165 SFRVFESY 172
> YIL050w
Length=285
Score = 81.3 bits (199), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 47/130 (36%), Positives = 67/130 (51%), Gaps = 11/130 (8%)
Query 18 CFLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVW-----------L 66
F P I++ YLER+ ++ + + F+ LVY DR + H +
Sbjct 149 AFYGKNVPEIAVVQYLERIQKYCPTTNDIFLSLLVYFDRISKNYGHSSERNGCAKQLFVM 208
Query 67 CPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMP 126
N+HRL +T + + KF D FYSN+ YAKVGG+ LQE+NHLE L + F+L V
Sbjct 209 DSGNIHRLLITGVTICTKFLSDFFYSNSRYAKVGGISLQELNHLELQFLILCDFKLLVSV 268
Query 127 CEFDKYFKLV 136
E KY L+
Sbjct 269 EEMQKYANLL 278
> SPBC20F10.10
Length=243
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 68/131 (51%), Gaps = 3/131 (2%)
Query 10 SSEGCGAPC--FLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQ-MNNHVWL 66
S PC F + P IS+ YL R+ ++ + + F+ L+Y+DR + + V++
Sbjct 61 SPTSLKNPCLIFSAKNVPSISIQAYLTRILKYCPATNDVFLSVLIYLDRIVHHFHFTVFI 120
Query 67 CPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMP 126
N+HR + A KF D FY+N+ YAKVGG+PL E+NHLE + F L +
Sbjct 121 NSFNIHRFLIAGFTAASKFFSDVFYTNSRYAKVGGIPLHELNHLELSFFVFNDFNLFISL 180
Query 127 CEFDKYFKLVL 137
+ Y L+L
Sbjct 181 EDLQAYGDLLL 191
> YER059w
Length=420
Score = 65.9 bits (159), Expect = 2e-11, Method: Composition-based stats.
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 0/67 (0%)
Query 70 NLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMPCEF 129
N+HRL + + V+ KF D FYSN+ Y++VGG+ LQE+NHLE L + F L + E
Sbjct 336 NIHRLIIAGITVSTKFLSDFFYSNSRYSRVGGISLQELNHLELQFLVLCDFELLISVNEL 395
Query 130 DKYFKLV 136
+Y L+
Sbjct 396 QRYADLL 402
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 25 PMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCP 68
P I + Y +R+ ++ + + F+ LVY DR + N V P
Sbjct 229 PQIGLDQYFQRIQKYCPTTNDVFLSLLVYFDRISKRCNSVTTTP 272
> YOL001w
Length=293
Score = 60.8 bits (146), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 38/120 (31%), Positives = 60/120 (50%), Gaps = 0/120 (0%)
Query 3 QHLGVQNSSEGCGAPCFLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNN 62
++ + S + + S P IS+ +Y RL +F + +L YID +
Sbjct 51 ENSATKKSDDQITLTRYHSKIPPNISIFNYFIRLTKFSSLEHCVLMTSLYYIDLLQTVYP 110
Query 63 HVWLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRL 122
L L HR +TA VA K D+F +NA+YAKVGG+ E+N LE L+ +++R+
Sbjct 111 DFTLNSLTAHRFLLTATTVATKGLCDSFSTNAHYAKVGGVRCHELNILENDFLKRVNYRI 170
> Hs21450735
Length=341
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 52/94 (55%), Gaps = 2/94 (2%)
Query 41 QCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKV- 99
Q + EC ++ LVY++R L + +CP N R+ + A+++A K DD N Y ++
Sbjct 183 QLTAECAIVTLVYLERLLTYA-EIDICPANWKRIVLGAILLASKVWDDQAVWNVDYCQIL 241
Query 100 GGLPLQEMNHLEATLLRMLHFRLHVMPCEFDKYF 133
+ +++MN LE L +L F ++V + KY+
Sbjct 242 KDITVEDMNELERQFLELLQFNINVPSSVYAKYY 275
> CE25684
Length=395
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/117 (30%), Positives = 55/117 (47%), Gaps = 8/117 (6%)
Query 24 EPMISMPD------YLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVT 77
E M PD ++ L Q + EC ++ LVYI+R L + LCP N R+ +
Sbjct 178 EQMTRDPDHRNIYRFVRNLFSSAQLTAECAIITLVYIERLLNYA-EMDLCPSNWRRVVLG 236
Query 78 ALMVAVKFADDTFYSNAYYAKV-GGLPLQEMNHLEATLLRMLHFRLHVMPCEFDKYF 133
++M+A K DD N Y ++ + +MN LE L L F + V + KY+
Sbjct 237 SIMLASKVWDDQAVWNVDYCQILRDTNVDDMNELERRFLECLDFNIEVPSSVYAKYY 293
> CE28185
Length=397
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/117 (30%), Positives = 55/117 (47%), Gaps = 8/117 (6%)
Query 24 EPMISMPD------YLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVT 77
E M PD ++ L Q + EC ++ LVYI+R L + LCP N R+ +
Sbjct 180 EQMTRDPDHRNIYRFVRNLFSSAQLTAECAIITLVYIERLLNYA-EMDLCPSNWRRVVLG 238
Query 78 ALMVAVKFADDTFYSNAYYAKV-GGLPLQEMNHLEATLLRMLHFRLHVMPCEFDKYF 133
++M+A K DD N Y ++ + +MN LE L L F + V + KY+
Sbjct 239 SIMLASKVWDDQAVWNVDYCQILRDTNVDDMNELERRFLECLDFNIEVPSSVYAKYY 295
> 7297875
Length=406
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 51/94 (54%), Gaps = 2/94 (2%)
Query 41 QCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKV- 99
Q + EC ++ LVY++R L + + P N R+ + A+++A K DD N Y ++
Sbjct 245 QLTAECAIITLVYLERLLTYA-ELDVGPCNWKRMVLGAILLASKVWDDQAVWNVDYCQIL 303
Query 100 GGLPLQEMNHLEATLLRMLHFRLHVMPCEFDKYF 133
+ +++MN LE L +L F ++V + KY+
Sbjct 304 KDITVEDMNELERQFLELLQFNINVPSSVYAKYY 337
> YGL134w
Length=433
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/104 (25%), Positives = 54/104 (51%), Gaps = 7/104 (6%)
Query 19 FLSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPL--QMNNHVWLCPLNL----- 71
F ++P +S D+L+R+ + +++A ID + N++ LNL
Sbjct 300 FYMKSKPTLSSADFLKRIQDKCEYQPTVYLVATFLIDTLFLTRDGNNILQLKLNLQEKEV 359
Query 72 HRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLL 115
HR+ + A+ ++ K +D +S+ Y++KV G+ + + LE +LL
Sbjct 360 HRMIIAAVRLSTKLLEDFVHSHEYFSKVCGISKRLLTKLEVSLL 403
> Hs22069238
Length=344
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 49/99 (49%), Gaps = 2/99 (2%)
Query 36 LARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVK-FADDTFYSNA 94
L + + + C ++ALVYI R L N + LCP N ++ + +++A K + + +S
Sbjct 83 LFQVIKLTAPCAIVALVYIKRLLTSAN-IDLCPTNWKKIVLGTMLLASKVWRNHGLWSVD 141
Query 95 YYAKVGGLPLQEMNHLEATLLRMLHFRLHVMPCEFDKYF 133
++ M+ +E L +L F +HV + KY+
Sbjct 142 DSQNSKDTAVENMSKMEKCFLELLEFNIHVSASVYAKYY 180
> Hs20472847_2
Length=301
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/115 (22%), Positives = 61/115 (53%), Gaps = 7/115 (6%)
Query 22 ATEPMISM--PDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTAL 79
A++P ++M ++ L + + + E A++YI+R + + + +CP N R+ + A+
Sbjct 127 ASQPHLTMTLKSFVRTLFKAMRLTAE---FAIIYIERLVSYAD-IDICPTNWKRIVLGAI 182
Query 80 MVAVKFADDTFYSNAYYAKV-GGLPLQEMNHLEATLLRMLHFRLHVMPCEFDKYF 133
++A K D N Y K+ + ++EMN LE L+++++ + + +++
Sbjct 183 LLASKVWSDMAVWNEDYCKLFKNITVEEMNELERQFLKLINYNNSITNSVYSRFY 237
> Hs17461133
Length=987
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 23/93 (24%), Positives = 46/93 (49%), Gaps = 1/93 (1%)
Query 41 QCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVG 100
Q + EC ++ LVY+++ L +CP N R+ + A++ + + ++ Y
Sbjct 853 QLTAECAIVTLVYLEKLLTYA-ETNICPTNWKRIGLRAILASKVWNHQAVWNMDYCQIFK 911
Query 101 GLPLQEMNHLEATLLRMLHFRLHVMPCEFDKYF 133
+ ++MN +E L +L F L+V + KY+
Sbjct 912 DITGEDMNEMERHFLMLLQFNLNVPASVYAKYY 944
> YPR120c
Length=435
Score = 37.0 bits (84), Expect = 0.010, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 45/94 (47%), Gaps = 5/94 (5%)
Query 31 DYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKFADDTF 90
D+L + FQC E L++ +DR L N V + L L LAVT+L +A KF +
Sbjct 203 DWLVEVHEKFQCYPETLFLSINLMDRFLA-KNKVTMNKLQL--LAVTSLFIAAKFEEVNL 259
Query 91 YSNAYYAKV--GGLPLQEMNHLEATLLRMLHFRL 122
A YA + G ++ + E +L L F +
Sbjct 260 PKLAEYAYITDGAASKNDIKNAEMFMLTSLEFNI 293
> SPAC19E9.03
Length=166
Score = 36.2 bits (82), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 72 HRLAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMPCEFDK 131
RL + LM A KF D +SN ++++ GLP+ ++ LE + + +RL V F +
Sbjct 84 RRLFLVCLMAATKFLQDRSFSNRAWSRLSGLPVDKLLVLEYMFYQCIDYRLVVPKHIFAR 143
Query 132 YFKLV 136
+ LV
Sbjct 144 WSLLV 148
> SPBC582.03
Length=482
Score = 35.4 bits (80), Expect = 0.031, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 46/97 (47%), Gaps = 7/97 (7%)
Query 29 MPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLN-LHRLAVTALMVAVKFAD 87
+ D+L + F+ E LA+ IDR L + +C LN L + + AL +A K+ +
Sbjct 239 LTDWLIEVHSRFRLLPETLFLAVNIIDRFLSLR----VCSLNKLQLVGIAALFIASKYEE 294
Query 88 DTFYS--NAYYAKVGGLPLQEMNHLEATLLRMLHFRL 122
S N Y GG +E+ E +LR+L F L
Sbjct 295 VMCPSVQNFVYMADGGYDEEEILQAERYILRVLEFNL 331
> CE09297
Length=374
Score = 35.4 bits (80), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 49/89 (55%), Gaps = 5/89 (5%)
Query 48 VLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKF--ADDTF--YSNAYYAKVGGLP 103
V+ALVY+DR L++ + + L V AL++A K+ DT+ SN+ +A+ +
Sbjct 125 VVALVYLDR-LRVQKKAFFESTDPVSLYVPALVLASKYMHDADTYDRVSNSEWAESLKMT 183
Query 104 LQEMNHLEATLLRMLHFRLHVMPCEFDKY 132
+++N E L++ L + + V EF+ Y
Sbjct 184 PEDLNAKEWNLVKNLDWNVAVKNDEFENY 212
> YNL289w
Length=279
Score = 35.0 bits (79), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 24/100 (24%), Positives = 53/100 (53%), Gaps = 3/100 (3%)
Query 28 SMPDYLERLARFFQCSGECFVLALVYIDRPLQ-MNNHVWLCPLNLHRLAVTALMVAVKFA 86
S+ ++ RL R+ + A Y+++ + + P +HR+ + L+++ KF
Sbjct 54 SLMTFITRLVRYTNVYTPTLLTAACYLNKLKRILPRDATGLPSTIHRIFLACLILSAKFH 113
Query 87 DDTFYSNAYYAKV--GGLPLQEMNHLEATLLRMLHFRLHV 124
+D+ N ++A+ G L+++N +E LL++L++ L V
Sbjct 114 NDSSPLNKHWARYTDGLFTLEDINLMERQLLQLLNWDLRV 153
> At1g77390
Length=454
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 3/57 (5%)
Query 31 DYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKFAD 87
D+L +A ++ S E LA+ Y+DR L N + NL L VT +M+A K+ +
Sbjct 217 DWLVEVAEEYRLSPETLYLAVNYVDRYLTGN---AINKQNLQLLGVTCMMIAAKYEE 270
> At5g43080
Length=355
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 47/100 (47%), Gaps = 17/100 (17%)
Query 31 DYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKFADDT- 89
D+L +A ++ + LA+ YIDR L + + L L VT++++A K+ + T
Sbjct 126 DWLVEVAEEYKLLSDTLYLAVSYIDRFLSLKT---VNKQRLQLLGVTSMLIASKYEEITP 182
Query 90 -------FYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRL 122
+ ++ Y K QE+ +EA +L L F L
Sbjct 183 PNVDDFCYITDNTYTK------QEIVKMEADILLALQFEL 216
> SPBC1D7.03c
Length=461
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 74 LAVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHVMP 126
L TAL++A KF DD ++N +++V G +N E L + + L P
Sbjct 336 LLTTALILANKFLDDNTFTNQSWSQVTGFRTALLNSFEQDWLASMSWNLSPGP 388
> At1g47220
Length=327
Score = 32.0 bits (71), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 42/96 (43%), Gaps = 9/96 (9%)
Query 31 DYLERLARFFQCSGECFVLALVYIDR--PLQMNNHVWLCPLNLHRLAVTALMVAVKFADD 88
D+L +A F+ E L + YIDR L+M N W L + V+A+ +A K+ +
Sbjct 97 DWLVEVAEEFELVSETLYLTVSYIDRFLSLKMVNEHW-----LQLVGVSAMFIASKYEEK 151
Query 89 TF--YSNAYYAKVGGLPLQEMNHLEATLLRMLHFRL 122
+ Y Q++ +E +L L F L
Sbjct 152 RRPKVEDFCYITANTYTKQDVLKMEEDILLALEFEL 187
> At5g65420
Length=308
Score = 31.2 bits (69), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 46/97 (47%), Gaps = 14/97 (14%)
Query 46 CFVLALVYIDRPLQMNN----HVWLCPLNLHRLAVTALMVAVKFADDTFYSNAYYAKVGG 101
CF LA+ Y+DR L +++ W+ L LAV L +A K ++T +VG
Sbjct 98 CFCLAMNYLDRFLSVHDLPSGKGWI----LQLLAVACLSLAAKI-EETEVPMLIDLQVGD 152
Query 102 ----LPLQEMNHLEATLLRMLHFRLH-VMPCEFDKYF 133
+ + +E +L L +RL + PC + +YF
Sbjct 153 PQFVFEAKSVQRMELLVLNKLKWRLRAITPCSYIRYF 189
> At1g29680
Length=237
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 25/45 (55%), Gaps = 4/45 (8%)
Query 20 LSATEPMISMPDYLERLARFFQCSGECFVLALVYIDRPLQMNNHV 64
L T I+ P+++ +A +++ G+CFVL + + +MN H
Sbjct 194 LKHTRAEIAEPEWINPMADYWKQHGKCFVLDIATV----EMNRHA 234
> CE21610
Length=405
Score = 29.6 bits (65), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 31 DYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKF 85
D++ +A+ C G+ F+LA+ IDR + + N + ++ +A AL +A K
Sbjct 110 DWIYDVAKEENCDGDVFLLAVSLIDRFMSVQN---ILKHDIQMIAGVALFIASKL 161
> At5g25380
Length=444
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 23/102 (22%), Positives = 48/102 (47%), Gaps = 17/102 (16%)
Query 31 DYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVKFADDT- 89
D+L ++ ++ + L + IDR + +H ++ L L +T +++A K+ + +
Sbjct 219 DWLVEVSEEYKLVSDTLYLTVNLIDRFM---SHNYIEKQKLQLLGITCMLIASKYEEISA 275
Query 90 -------FYSNAYYAKVGGLPLQEMNHLEATLLRMLHFRLHV 124
F ++ Y ++ E+ +E +L LHFRL V
Sbjct 276 PRLEEFCFITDNTYTRL------EVLSMEIKVLNSLHFRLSV 311
> Hs18591298
Length=753
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 19/80 (23%), Positives = 37/80 (46%), Gaps = 10/80 (12%)
Query 55 DRPLQMNNHVWLCPLNLHRLAVTALMVAVK-FADDTFYSNAYYAKVGGLPLQEMNHLEAT 113
DRPL P L + + A+++ K + D T ++ Y +++MN LE
Sbjct 57 DRPL---------PDQLEEIVMGAILLTSKVWKDVTIWNREYCRLFVNTSIKDMNELERQ 107
Query 114 LLRMLHFRLHVMPCEFDKYF 133
L+++ + + V + KY+
Sbjct 108 FLQLIDYNVEVSGSVYAKYY 127
> 7298245
Length=709
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 2/54 (3%)
Query 31 DYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLNLHRLAVTALMVAVK 84
D+L + ++ E F LA+ Y+DR L + + V L L + +T L VA K
Sbjct 367 DWLIEVCEVYKLHRETFYLAVDYLDRYLHVAHKVQKTHLQL--IGITCLFVAAK 418
> YHR190w
Length=444
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 75 AVTALMVAVKFADDTFYSNAYYAKVGGLPLQEMN 108
+T L+V KFA+++ YSN + GL LQ+ N
Sbjct 189 GLTRLIVIAKFANESLYSNEQLYESMGLFLQKTN 222
> YGR109c
Length=380
Score = 27.3 bits (59), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 41/95 (43%), Gaps = 7/95 (7%)
Query 31 DYLERLARFFQCSGECFVLALVYIDRPLQMNNHVWLCPLN-LHRLAVTALMVAVKFADDT 89
D+L + F C E LA+ +DR L N + LN L L +T L +A KF +
Sbjct 161 DWLVEVHEKFHCLPETLFLAINLLDRFLSQN----VVKLNKLQLLCITCLFIACKFEEVK 216
Query 90 F--YSNAYYAKVGGLPLQEMNHLEATLLRMLHFRL 122
+N Y G ++ + E +L L + +
Sbjct 217 LPKITNFAYVTDGAATVEGIRKAELFVLSSLGYNI 251
Lambda K H
0.330 0.142 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40