bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5827_orf2
Length=109
Score E
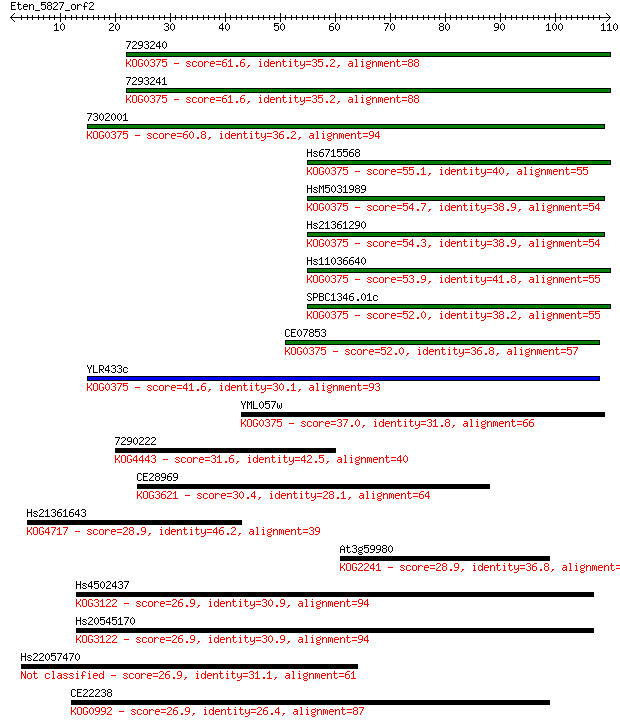
Sequences producing significant alignments: (Bits) Value
7293240 61.6 3e-10
7293241 61.6 3e-10
7302001 60.8 7e-10
Hs6715568 55.1 3e-08
HsM5031989 54.7 4e-08
Hs21361290 54.3 5e-08
Hs11036640 53.9 6e-08
SPBC1346.01c 52.0 3e-07
CE07853 52.0 3e-07
YLR433c 41.6 4e-04
YML057w 37.0 0.008
7290222 31.6 0.42
CE28969 30.4 0.87
Hs21361643 28.9 2.3
At3g59980 28.9 2.5
Hs4502437 26.9 8.3
Hs20545170 26.9 9.3
Hs22057470 26.9 9.6
CE22238 26.9 10.0
> 7293240
Length=570
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 31/88 (35%), Positives = 53/88 (60%), Gaps = 2/88 (2%)
Query 22 ELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENE 81
E+ V+ I + ++ EE P S R+ E +R K++++G++ RVF +REE+E
Sbjct 429 EMLVNVLNICSDDELMTEESEEPLSDDEAALRK--EVIRNKIRAIGKMARVFSVLREESE 486
Query 82 LIMQLKGCTPGDRIPVGLLIQGREGLEN 109
++QLKG TP +P+G L G++ L+N
Sbjct 487 SVLQLKGLTPTGALPLGALSGGKQSLKN 514
> 7293241
Length=575
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 31/88 (35%), Positives = 53/88 (60%), Gaps = 2/88 (2%)
Query 22 ELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENE 81
E+ V+ I + ++ EE P S R+ E +R K++++G++ RVF +REE+E
Sbjct 437 EMLVNVLNICSDDELMTEESEEPLSDDEAALRK--EVIRNKIRAIGKMARVFSVLREESE 494
Query 82 LIMQLKGCTPGDRIPVGLLIQGREGLEN 109
++QLKG TP +P+G L G++ L+N
Sbjct 495 SVLQLKGLTPTGALPLGALSGGKQSLKN 522
> 7302001
Length=568
Score = 60.8 bits (146), Expect = 7e-10, Method: Composition-based stats.
Identities = 34/94 (36%), Positives = 56/94 (59%), Gaps = 2/94 (2%)
Query 15 DEEIDNVELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFK 74
D+E++ EL ++V ++ AN S +S R E +R K++++G++ RVF
Sbjct 368 DDELEE-ELRKKIV-LVPANASNNNNNNNTPSKPASMSALRKEIIRNKIRAIGKMSRVFS 425
Query 75 TIREENELIMQLKGCTPGDRIPVGLLIQGREGLE 108
+REE+E ++QLKG TP +PVG L GR+ L+
Sbjct 426 ILREESESVLQLKGLTPTGALPVGALSGGRDSLK 459
> Hs6715568
Length=521
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ RVF +REE+E ++ LKG TP +P G+L G++ L++
Sbjct 392 RKEVIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLPSGVLSGGKQTLQS 446
> HsM5031989
Length=502
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLE 108
R E +R K++++G++ RVF +R+E+E ++ LKG TP +P+G+L G++ +E
Sbjct 386 RKEIIRNKIRAIGKMARVFSILRQESESVLTLKGLTPTGTLPLGVLSGGKQTIE 439
> Hs21361290
Length=512
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLE 108
R E +R K++++G++ RVF +R+E+E ++ LKG TP +P+G+L G++ +E
Sbjct 386 RKEIIRNKIRAIGKMARVFSILRQESESVLTLKGLTPTGTLPLGVLSGGKQTIE 439
> Hs11036640
Length=524
Score = 53.9 bits (128), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ RVF +REE+E ++ LKG TP +P G+L GR+ L++
Sbjct 401 RKEIIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLPSGVLAGGRQTLQS 455
> SPBC1346.01c
Length=554
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R + ++ K+ ++GR+ RVF +REE E + +LK + R+P G L+ G EG++N
Sbjct 440 RRQIIKNKIMAIGRISRVFSVLREERESVSELKNVSGTQRLPAGTLMLGAEGIKN 494
> CE07853
Length=535
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 51 ISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGL 107
+ R E +R K++++G++ R F +REE+E ++ LKG TP +P+G L G G+
Sbjct 414 VGSARKEVIRHKIRAIGKMARAFSVLREESESVLALKGLTPTGALPMGTLQGGSRGV 470
> YLR433c
Length=553
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 48/97 (49%), Gaps = 5/97 (5%)
Query 15 DEEIDNVELPPEVVKIMRANIVLAEEGTAPR----SAGTPISRERAEALRRKVQSVGRLM 70
++E+D P + ++A +E P S+ R +ALR K+ ++ ++
Sbjct 410 EQELDPESEPKAAEETVKARANATKETGTPSDEKASSAILEDETRRKALRNKILAIAKVS 469
Query 71 RVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGL 107
R+F +REE+E + LK G +P G L +G EGL
Sbjct 470 RMFSVLREESEKVEYLKTMNAG-VLPRGALARGTEGL 505
> YML057w
Length=604
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 35/66 (53%), Gaps = 1/66 (1%)
Query 43 APRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQ 102
+P+ A R +ALR K+ +V ++ R++ +REE + LK G +P G L
Sbjct 489 SPKHASILDDEHRRKALRNKILAVAKVSRMYSVLREETNKVQFLKDHNSG-VLPRGALSN 547
Query 103 GREGLE 108
G +GL+
Sbjct 548 GVKGLD 553
> 7290222
Length=2304
Score = 31.6 bits (70), Expect = 0.42, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query 20 NVELPPEVVKI------MRANIVLAEEGTAPRSAGTPISRERAEAL 59
+V L P+VVK+ +A +++ +E S GTP S ERAE +
Sbjct 565 SVSLAPDVVKLEEVGSESKAKLLVKQEAVVKDSTGTPTSEERAEEI 610
> CE28969
Length=1125
Score = 30.4 bits (67), Expect = 0.87, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 24 PPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELI 83
PP++VK++R L T P + TPI+ A A V ++G + + R+++E+
Sbjct 644 PPQIVKVIRPGHRLQTTTTKPIATVTPITVPVANADDVAVNNMGMISEEMR--RKQDEMW 701
Query 84 MQLK 87
+ L+
Sbjct 702 LDLR 705
> Hs21361643
Length=765
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 4/40 (10%)
Query 4 ILNPSLEEGEEDEEIDNVE-LPPEVVKIMRANIVLAEEGT 42
+LN EEGE D+E D E LPP K+ R + +A GT
Sbjct 485 VLNQIFEEGESDDEFDMDENLPP---KLSRLKMNIASPGT 521
> At3g59980
Length=273
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 61 RKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVG 98
R V+S G L+ E EL++ +G PGDR+ G
Sbjct 173 RGVKSCGMLLAASDAAHENVELLVPPEGSVPGDRVWFG 210
> Hs4502437
Length=395
Score = 26.9 bits (58), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 42/99 (42%), Gaps = 14/99 (14%)
Query 13 EEDEEIDNVELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRV 72
EED E+D +P VK + + AP P + + L + V SV L+R
Sbjct 204 EEDMEVD---IPAVKVKEEPRDEEEEAKMKAP-----PKAARKTPGLPKDV-SVAELLRE 254
Query 73 FKTIREENELIMQLKGCTPG-----DRIPVGLLIQGREG 106
+EE L +QL PG D P+ +QG +G
Sbjct 255 LSLTKEEELLFLQLPDTLPGQPPTQDIKPIKTEVQGEDG 293
> Hs20545170
Length=398
Score = 26.9 bits (58), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 42/99 (42%), Gaps = 14/99 (14%)
Query 13 EEDEEIDNVELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRV 72
EED E+D +P VK + + AP P + + L + V SV L+R
Sbjct 207 EEDMEVD---IPAVKVKEEPRDEEEEAKMKAP-----PKAARKTPGLPKDV-SVAELLRE 257
Query 73 FKTIREENELIMQLKGCTPG-----DRIPVGLLIQGREG 106
+EE L +QL PG D P+ +QG +G
Sbjct 258 LSLTKEEELLFLQLPDTLPGQPPTQDIKPIKTEVQGEDG 296
> Hs22057470
Length=754
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 15/61 (24%)
Query 3 GILNPSLEEGEEDEEIDNVELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRK 62
G+ +PSL E E +EE+ + +RAN +L G I+ + E++RRK
Sbjct 675 GLKSPSLSEAEIEEELSS------AANSIRANRLLTTRG---------IASQEEESVRRK 719
Query 63 V 63
V
Sbjct 720 V 720
> CE22238
Length=734
Score = 26.9 bits (58), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 43/87 (49%), Gaps = 3/87 (3%)
Query 12 GEEDEEIDNVELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMR 71
GEE + I+N E + +IM IV+ + A R+ E ++Q ++++
Sbjct 572 GEEAKRIENEEQKLNMQQIMIDKIVILQRKLARRTEKCEFLEEHVRQCLEELQKKTKIIQ 631
Query 72 VFKTIREENELIMQLKGCTPGDRIPVG 98
F +REE L+M +G +++P+G
Sbjct 632 HF-ALREEASLLMPSEGSL--EKVPIG 655
Lambda K H
0.314 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1199474506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40