bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5838_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
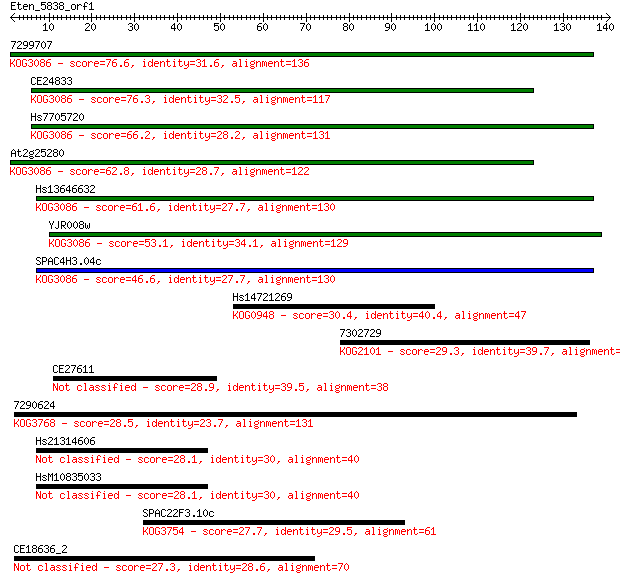
7299707 76.6 1e-14
CE24833 76.3 2e-14
Hs7705720 66.2 2e-11
At2g25280 62.8 2e-10
Hs13646632 61.6 5e-10
YJR008w 53.1 1e-07
SPAC4H3.04c 46.6 2e-05
Hs14721269 30.4 1.1
7302729 29.3 2.5
CE27611 28.9 3.1
7290624 28.5 4.0
Hs21314606 28.1 5.1
HsM10835033 28.1 5.4
SPAC22F3.10c 27.7 7.2
CE18636_2 27.3 8.4
> 7299707
Length=295
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/136 (31%), Positives = 71/136 (52%), Gaps = 7/136 (5%)
Query 1 EGLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREA 60
E + Y L + MDP LFVIS+D+ HWG R SYTY + ++K I+ LD++
Sbjct 160 EQEAQYGSLLSSYLMDPTNLFVISSDFCHWGHRFSYTYYDSSCG---AIHKSIEKLDKQG 216
Query 61 IDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQ 120
+D + +LN E++ K N ICG + + L ++ + + + D S L Y+Q
Sbjct 217 MDIIESLNPHSFTEYLRKYNNTICGRHPIGVMLGAVKALQD----QGYDKMSFKFLKYAQ 272
Query 121 SSLIRSASENAVGYVA 136
SS + +++V Y +
Sbjct 273 SSQCQDIEDSSVSYAS 288
> CE24833
Length=392
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/117 (32%), Positives = 64/117 (54%), Gaps = 6/117 (5%)
Query 6 YAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVV 65
Y + DP LFVIS+D+ HWG R S++ P + S+P+Y++I +D++ + +
Sbjct 260 YGNIFAHYMEDPRNLFVISSDFCHWGERFSFS--PYDRHSSIPIYEQITNMDKQGMSAIE 317
Query 66 NLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSS 122
LN + +++ KT N ICG + +L+ L+ E S+ E F L Y+QS+
Sbjct 318 TLNPAAFNDYLKKTQNTICGRNPILIMLQAAEHFRISNNHTHEFRF----LHYTQSN 370
> Hs7705720
Length=297
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/131 (28%), Positives = 67/131 (51%), Gaps = 8/131 (6%)
Query 6 YAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVV 65
+ + + DP LFV+S+D+ HWG R Y+Y E+ + +Y+ I+ LD+ + +
Sbjct 169 FGKLFSKYLADPSNLFVVSSDFCHWGQRFRYSYYDESQGE---IYRSIEHLDKMGMSIIE 225
Query 66 NLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSSLIR 125
L+ ++ K N ICG + + L + + Q++ S S L+Y+QSS R
Sbjct 226 QLDPVSFSNYLKKYHNTICGRHPIGVLLNAITEL-----QKNGMNMSFSFLNYAQSSQCR 280
Query 126 SASENAVGYVA 136
+ +++V Y A
Sbjct 281 NWQDSSVSYAA 291
> At2g25280
Length=291
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 62/122 (50%), Gaps = 11/122 (9%)
Query 1 EGLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREA 60
E +MY E L + DP F +S+D+ HWG R +Y + +N + ++K I+ALD++
Sbjct 160 ENEAMYGELLAKYVDDPKNFFSVSSDFCHWGSRFNYMHY-DNTHGA--IHKSIEALDKKG 216
Query 61 IDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQ 120
+D + + ++++ + N ICG + +FL +L + S + L Y Q
Sbjct 217 MDIIETGDPDAFKKYLLEFENTICGRHPISIFLHML--------KHSSSKIKINFLRYEQ 268
Query 121 SS 122
SS
Sbjct 269 SS 270
> Hs13646632
Length=295
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 36/130 (27%), Positives = 66/130 (50%), Gaps = 8/130 (6%)
Query 7 AETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVVN 66
+E + DP LFV+S+D+ HWG R ++Y E+ + + + I+ LD+ + +
Sbjct 168 SELFSKYLADPSNLFVVSSDFCHWGQRFRHSYYDESQGE---ICRSIEHLDKMGMSIIEQ 224
Query 67 LNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSSLIRS 126
L+ ++ K N ICG + + L + + Q++ S S L+Y+QSS R+
Sbjct 225 LDPVSFSNYLKKYHNTICGRHPIGVLLNAITEL-----QKNGMNMSFSFLNYAQSSQCRN 279
Query 127 ASENAVGYVA 136
+++V Y A
Sbjct 280 WQDSSVSYAA 289
> YJR008w
Length=338
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 44/162 (27%), Positives = 76/162 (46%), Gaps = 39/162 (24%)
Query 10 LVPFFMDPDTLFVISTDWSHWGPRSSYT-----------YLPENVD------------KS 46
L + DP+ LF++S+D+ HWG R YT + E +
Sbjct 183 LSEYIKDPNNLFIVSSDFCHWGRRFQYTGYVGSKEELNDAIQEETEVEMLTARSKLSHHQ 242
Query 47 LPLYKKIQALDREAIDCVVNL-NGS---CLEEHVAKTGNRICGYDALLLFLRLLERV--A 100
+P+++ I+ +DR A+ + + NG ++++ TGN ICG + + L L ++ A
Sbjct 243 VPIWQSIEIMDRYAMKTLSDTPNGERYDAWKQYLEITGNTICGEKPISVILSALSKIRDA 302
Query 101 ESSGQRSEDLFSASLLSYSQSSLIRSASENAV----GYVAIG 138
SG + + +YSQSS + S +++V GYV IG
Sbjct 303 GPSGIKFQ------WPNYSQSSHVTSIDDSSVSYASGYVTIG 338
> SPAC4H3.04c
Length=309
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/145 (24%), Positives = 61/145 (42%), Gaps = 23/145 (15%)
Query 7 AETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDK---------------SLPLYK 51
A+ L + D FVIS+D+ HWG R YT + ++ S +Y+
Sbjct 170 AKFLSQYIKDESNSFVISSDFCHWGRRFGYTLYLNDTNQLEDAVLKYKRRGGPTSPKIYE 229
Query 52 KIQALDREAIDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLF 111
I LD + + + E++ T N ICG + L ++ +E SE
Sbjct 230 SISNLDHIGMKIIETKSSDDFSEYLKTTQNTICGRYPIELIMKSME-----CANFSERF- 283
Query 112 SASLLSYSQSSLIRSASENAVGYVA 136
+SY+QSS + ++++V Y
Sbjct 284 --KFISYAQSSHVELVTDSSVSYAT 306
> Hs14721269
Length=1042
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query 53 IQALDREAIDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERV 99
+ A REAI CV N N S L G +C A+ L LR +RV
Sbjct 140 LDAFQREAIQCVDN-NQSVLVSAHTSAGKTVCAEYAIALALREKQRV 185
> 7302729
Length=407
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 8/60 (13%)
Query 78 KTGNRICGYDALLLFL--RLLERVAESSGQRSEDLFSASLLSYSQSSLIRSASENAVGYV 135
KTGN +CG D F +LL R S S DL + Y +SL+RS SE V +
Sbjct 50 KTGNSVCGLDKRSQFREQKLLGRKCHS----SPDL--RQIGKYQNNSLLRSKSEGDVSLI 103
> CE27611
Length=604
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 2/38 (5%)
Query 11 VPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLP 48
VPF P TL D S W S+Y P VD + P
Sbjct 304 VPF--APGTLPTTKVDASKWNDPSAYATAPSTVDSTTP 339
> 7290624
Length=1194
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 31/140 (22%), Positives = 59/140 (42%), Gaps = 24/140 (17%)
Query 2 GLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQAL----- 56
G+ Y + PF+++P + VI TD + R+ + + LPL +I
Sbjct 66 GIDTYGQGRCPFYLEPSVIIVI-TDGGRYSYRNGV-----HQEIILPLSNQIPGTKFTKE 119
Query 57 ----DREAIDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFS 112
D+ V+ + G+ ++E V ++ D+ +ER+ E +G RS + S
Sbjct 120 PFRWDQRLFSLVLRMPGNKIDERV---DGKVPHDDS------PIERMCEVTGGRSYRVRS 170
Query 113 ASLLSYSQSSLIRSASENAV 132
+L+ SL++ V
Sbjct 171 HYVLNQCIESLVQKVQPGVV 190
> Hs21314606
Length=511
Score = 28.1 bits (61), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 7 AETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKS 46
A+ + F+M D L + DW W P + + P++V K+
Sbjct 127 AKKDITFYMPVDNLGMAVIDWEEWRPTWARNWKPKDVYKN 166
> HsM10835033
Length=509
Score = 28.1 bits (61), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 7 AETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKS 46
A+ + F+M D L + DW W P + + P++V K+
Sbjct 127 AKKDITFYMPVDNLGMAVIDWEEWRPTWARNWKPKDVYKN 166
> SPAC22F3.10c
Length=669
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 5/64 (7%)
Query 32 PRSSYTYLP---ENVDKSLPLYKKIQALDREAIDCVVNLNGSCLEEHVAKTGNRICGYDA 88
P+S Y + N ++LP Y + + + DC L C++E +AK I D
Sbjct 379 PKSRYDSVDLYISNDKRNLPEYNDVPVVINQ--DCYDKLIKDCIDERLAKHMAHIFSRDP 436
Query 89 LLLF 92
L++F
Sbjct 437 LVIF 440
> CE18636_2
Length=2051
Score = 27.3 bits (59), Expect = 8.4, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 32/72 (44%), Gaps = 13/72 (18%)
Query 2 GLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAI 61
GL Y + ++P TDWS RS+ + +DK+L K Q+ + AI
Sbjct 1508 GLQKYFQIILPV-----------TDWSIESNRSALNIILRRLDKTLSKIAKRQSFRKRAI 1556
Query 62 DCVVN--LNGSC 71
++ +NG C
Sbjct 1557 WIALSSWINGIC 1568
Lambda K H
0.319 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1571531578
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40