bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5840_orf4
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
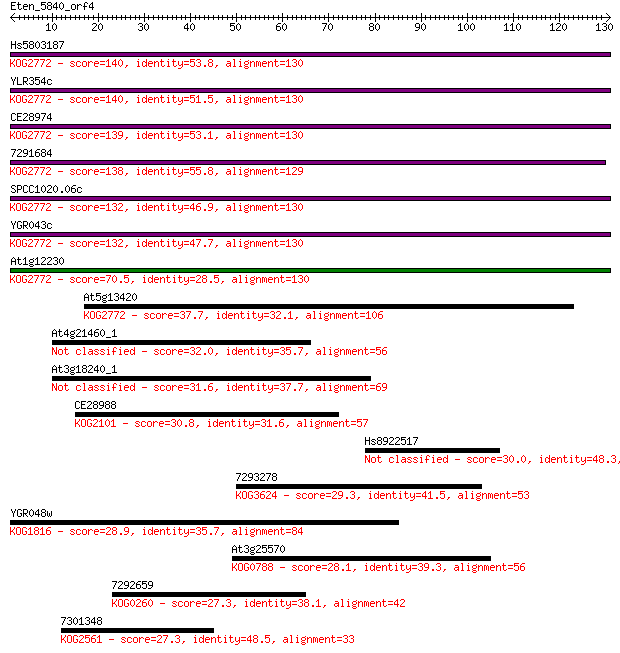
Hs5803187 140 4e-34
YLR354c 140 7e-34
CE28974 139 2e-33
7291684 138 2e-33
SPCC1020.06c 132 2e-31
YGR043c 132 2e-31
At1g12230 70.5 8e-13
At5g13420 37.7 0.006
At4g21460_1 32.0 0.33
At3g18240_1 31.6 0.37
CE28988 30.8 0.69
Hs8922517 30.0 1.2
7293278 29.3 2.0
YGR048w 28.9 2.3
At3g25570 28.1 4.5
7292659 27.3 7.7
7301348 27.3 8.1
> Hs5803187
Length=337
Score = 140 bits (354), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 70/130 (53%), Positives = 92/130 (70%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF AI E+ P DATTNP+L+L A Y+ L ++AI ++ G E ++ A+
Sbjct 27 DTGDFHAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGRKL--GGSQEDQIKNAI 84
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++ V FG E+L+ IPG VSTE+ A LSFD + VARARR+IELYK GISKDRILIK++
Sbjct 85 DKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYKEAGISKDRILIKLS 144
Query 121 ATWEGIQAAK 130
+TWEGIQA K
Sbjct 145 STWEGIQAGK 154
> YLR354c
Length=335
Score = 140 bits (352), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 67/130 (51%), Positives = 94/130 (72%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF +I +F P D+TTNP+L+L A Y L A+ ++ GK E VE AV
Sbjct 29 DTGDFGSIAKFQPQDSTTNPSLILAAAKQPTYAKLIDVAVEYGKK--HGKTTEEQVENAV 86
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
DR+LV FG E+L+I+PG VSTE+ A LSFDT+ ++ +AR II+L++ +G+SK+R+LIK+A
Sbjct 87 DRLLVEFGKEILKIVPGRVSTEVDARLSFDTQATIEKARHIIKLFEQEGVSKERVLIKIA 146
Query 121 ATWEGIQAAK 130
+TWEGIQAAK
Sbjct 147 STWEGIQAAK 156
> CE28974
Length=319
Score = 139 bits (349), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 69/130 (53%), Positives = 94/130 (72%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF AIKEF PTDATTNP+L+L A +Y L +++ A+E + G + ++ A+
Sbjct 16 DTGDFNAIKEFQPTDATTNPSLILAASKMEQYAALIDQSVAYAKEHASGH--QEVLQAAM 73
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
DR+ V FG E+L+ IPG VSTE+ A LSFDT+ S+ RA +I Y+ +GISKDRILIK+A
Sbjct 74 DRLFVVFGKEILKTIPGRVSTEVDARLSFDTQASIDRALGLIAQYEKEGISKDRILIKLA 133
Query 121 ATWEGIQAAK 130
+TWEGI+AAK
Sbjct 134 STWEGIRAAK 143
> 7291684
Length=320
Score = 138 bits (348), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 72/131 (54%), Positives = 96/131 (73%), Gaps = 6/131 (4%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGK--DLEATVEE 58
D+ DFEAI + PTDATTNP+L+L A S +Y+ L QKA+ E +KGK + V E
Sbjct 16 DTGDFEAINIYKPTDATTNPSLILSASSMERYQPLVQKAV----EYAKGKKGSVSEQVAE 71
Query 59 AVDRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIK 118
A+D + V FG E+L+++PG VSTEI A LSFDT+ SV +A ++I LYKS G+ K+RILIK
Sbjct 72 AMDYLCVLFGTEILKVVPGRVSTEIDARLSFDTKKSVEKALKLIALYKSLGVDKERILIK 131
Query 119 VAATWEGIQAA 129
+A+TWEGI+AA
Sbjct 132 LASTWEGIKAA 142
> SPCC1020.06c
Length=322
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 61/130 (46%), Positives = 96/130 (73%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DFE+I ++ P DATTNP+L+L A +Y L A++ A+ +KG + + +E A
Sbjct 17 DTGDFESIAKYKPQDATTNPSLILAASKKPQYAALVDAAVDYAK--AKGGSINSQIEIAF 74
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
DR+L+ FG ++L I+PG VSTE+ A SFDT+ ++ +AR +I+LY+++GI ++R+LIK+A
Sbjct 75 DRLLIEFGTKILAIVPGRVSTEVDARYSFDTQTTIEKARHLIKLYEAEGIGRERVLIKIA 134
Query 121 ATWEGIQAAK 130
+T+EGIQAAK
Sbjct 135 STYEGIQAAK 144
> YGR043c
Length=333
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 62/130 (47%), Positives = 90/130 (69%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
DS DFEAI ++ P D+TTNP+L+L A KY A+ ++ GK +E A+
Sbjct 29 DSGDFEAISKYEPQDSTTNPSLILAASKLEKYARFIDAAVEYGRK--HGKTDHEKIENAM 86
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++LV FG ++L+++PG VSTE+ A LSFD + +V +A II+LYK G+ K+R+LIK+A
Sbjct 87 DKILVEFGTQILKVVPGRVSTEVDARLSFDKKATVKKALHIIKLYKDAGVPKERVLIKIA 146
Query 121 ATWEGIQAAK 130
+TWEGIQAA+
Sbjct 147 STWEGIQAAR 156
> At1g12230
Length=377
Score = 70.5 bits (171), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 64/130 (49%), Gaps = 27/130 (20%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ F+ + F PT AT + L+L + + +
Sbjct 83 DTVVFDDFERFPPTAATVSSALLLGICG---------------------------LPDTI 115
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
R +V G +L++++PG VSTE+ A L++DT G + + ++ LY + DR+L K+
Sbjct 116 FRAIVNVGGDLVKLVPGRVSTEVDARLAYDTNGIIRKVHDLLRLYNEIDVPHDRLLFKIP 175
Query 121 ATWEGIQAAK 130
ATW+GI+AA+
Sbjct 176 ATWQGIEAAR 185
> At5g13420
Length=438
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 56/114 (49%), Gaps = 20/114 (17%)
Query 17 TTNPTLVLQAVSNSK-YKNLFQKAINEAQEASKGKDLEATVEEAVDRVLVGFGMELLEII 75
T+NP + +A+S S Y + F+ + GKD+E+ E V + + +L E I
Sbjct 112 TSNPAIFQKAISTSNAYNDQFRTLVES------GKDIESAYWELVVKDIQD-ACKLFEPI 164
Query 76 -------PGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVAAT 122
G VS E+ L+ DT+G+V A+ Y SK +++ + IK+ AT
Sbjct 165 YDQTEGADGYVSVEVSPRLADDTQGTVEAAK-----YLSKVVNRRNVYIKIPAT 213
> At4g21460_1
Length=280
Score = 32.0 bits (71), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 30/56 (53%), Gaps = 4/56 (7%)
Query 10 EFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAVDRVLV 65
E SPTD++ LV++ VSN + K+ K+ NE E + L +E + R LV
Sbjct 61 ESSPTDSSDKKDLVVKDVSNKELKSRIDKSFNEGNEDA----LPGVIEALLQRRLV 112
> At3g18240_1
Length=279
Score = 31.6 bits (70), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 35/73 (47%), Gaps = 8/73 (10%)
Query 10 EFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAVDRVLVGFGM 69
E SPTD+ LV++ VSN + K+ +K NE E + L +E + R LV
Sbjct 60 ESSPTDSPEKKDLVVEDVSNKELKSRIEKYFNEGNEDA----LPGVIEALLQRRLVDKHA 115
Query 70 ----ELLEIIPGL 78
ELLE I L
Sbjct 116 ETDDELLEKIESL 128
> CE28988
Length=994
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 15 DATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAVDRVLVGFGMEL 71
DATT+ +L AV +S Y N F E+ + +D+ A D +L+ F L
Sbjct 893 DATTSYSLATNAVYSSVYGNNFDGVFKESCRSLGDEDMRLWTRSAYDSILLLFWQPL 949
> Hs8922517
Length=557
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 78 LVSTEIPAELSFDTEGSVARARRIIELYK 106
LVS E LSF EG+++RAR+I L++
Sbjct 203 LVSREAVVALSFLIEGTISRARKIYPLHE 231
> 7293278
Length=879
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 10/63 (15%)
Query 50 KDLEATVEEAVDRVLVGF--GMELLEIIP--------GLVSTEIPAELSFDTEGSVARAR 99
KDLEA ++A++ V G G I P GL + +P SFD S++R R
Sbjct 67 KDLEAARQDALNEVAAGGAGGRTPAAITPNSTHPSANGLRALLLPGRRSFDALMSLSRKR 126
Query 100 RII 102
RI+
Sbjct 127 RIL 129
> YGR048w
Length=361
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 13/89 (14%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D A+F K F P A + +++ N +Y LF+ NE + G LE EE
Sbjct 40 DDANFGG-KIFLPPSALSKLSML-----NIRYPMLFKLTANETGRVTHGGVLEFIAEEG- 92
Query 61 DRV-LVGFGMELLEIIPG----LVSTEIP 84
RV L + ME L I PG + ST++P
Sbjct 93 -RVYLPQWMMETLGIQPGSLLQISSTDVP 120
> At3g25570
Length=349
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 32/59 (54%), Gaps = 3/59 (5%)
Query 49 GKDLEA-TVEEAVDRVLVGFGMELLEI-IPGLVSTEIPA-ELSFDTEGSVARARRIIEL 104
G DL+A +E VDRVLV FG E + + + TE+ A + D G ++ R + EL
Sbjct 264 GYDLKALNFKELVDRVLVCFGPEEFSVAVHANLGTEVLASDCVADVNGYFSQERELEEL 322
> 7292659
Length=1887
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 2/44 (4%)
Query 23 VLQAVSNSKYK--NLFQKAINEAQEASKGKDLEATVEEAVDRVL 64
+ QA+ +K N+ QKA N E + G L T E V+R+L
Sbjct 694 IQQAIKKAKDDVINVIQKAHNMELEPTPGNTLRQTFENKVNRIL 737
> 7301348
Length=614
Score = 27.3 bits (59), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Query 12 SPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQ 44
S T+AT N T L +SNSK NL Q ++EA+
Sbjct 539 SSTNATANSTTPLVNLSNSKV-NLLQTVLSEAK 570
Lambda K H
0.312 0.129 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40