bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5888_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
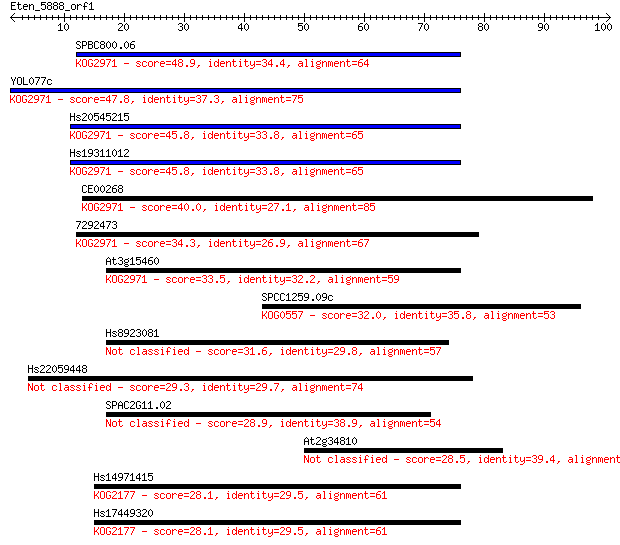
SPBC800.06 48.9 2e-06
YOL077c 47.8 5e-06
Hs20545215 45.8 2e-05
Hs19311012 45.8 2e-05
CE00268 40.0 0.001
7292473 34.3 0.062
At3g15460 33.5 0.11
SPCC1259.09c 32.0 0.26
Hs8923081 31.6 0.33
Hs22059448 29.3 1.9
SPAC2G11.02 28.9 2.2
At2g34810 28.5 3.1
Hs14971415 28.1 4.3
Hs17449320 28.1 4.5
> SPBC800.06
Length=295
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 41/67 (61%), Gaps = 3/67 (4%)
Query 12 QTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKVPLGPT--FIARLMNGLE 68
+K + K++L + EL +L +CN++ + E +R+ D LH+A+ P GPT F ++ ++
Sbjct 74 DSKLDSKDRLYQLNELAELYNCNNIFFFESRRREDLYLHIARAPNGPTVKFHVENLHTMD 133
Query 69 VLRFSGN 75
L +GN
Sbjct 134 ELNMTGN 140
> YOL077c
Length=291
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 48/82 (58%), Gaps = 7/82 (8%)
Query 1 HFVWALGFVLSQT----KWEKKEKLQSIAELCQLRSCNSVLYLEQRKN-DALLHVAKVPL 55
H + L +L + K + K+ LQ + E+ +L +CN+VL+ E RK+ D L ++K P
Sbjct 47 HLIQDLSGLLPHSRKEPKLDTKKDLQQLNEIAELYNCNNVLFFEARKHQDLYLWLSKPPN 106
Query 56 GPT--FIARLMNGLEVLRFSGN 75
GPT F + ++ ++ L F+GN
Sbjct 107 GPTIKFYIQNLHTMDELNFTGN 128
> Hs20545215
Length=353
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 44/68 (64%), Gaps = 3/68 (4%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKVPLGPT--FIARLMNGL 67
+ TK ++K+KL I E+C++++CN +Y E ++K D + ++ P GP+ F+ + ++ L
Sbjct 90 ADTKMDRKDKLFVINEVCEMKNCNKCIYFEAKKKQDLYMWLSNSPHGPSAKFLVQNIHTL 149
Query 68 EVLRFSGN 75
L+ +GN
Sbjct 150 AELKMTGN 157
> Hs19311012
Length=354
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 44/68 (64%), Gaps = 3/68 (4%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKVPLGPT--FIARLMNGL 67
+ TK ++K+KL I E+C++++CN +Y E ++K D + ++ P GP+ F+ + ++ L
Sbjct 90 ADTKMDRKDKLFVINEVCEMKNCNKCIYFEAKKKQDLYMWLSNSPHGPSAKFLVQNIHTL 149
Query 68 EVLRFSGN 75
L+ +GN
Sbjct 150 AELKMTGN 157
> CE00268
Length=352
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 48/89 (53%), Gaps = 4/89 (4%)
Query 13 TKWEKKEKLQSIAELCQLRSCNSVLYLEQRK-NDALLHVAKVPLGPT--FIARLMNGLEV 69
+K ++++ L + E+ ++++C V+Y E RK D L ++ V GP+ F+ ++ ++
Sbjct 95 SKLDQQKSLNVLNEIAEMKNCTKVMYFESRKRKDTYLWMSNVEKGPSIKFLVHNVHTMKE 154
Query 70 LRFSGNTER-TPPVGKLSAALSASANIRI 97
L+ SGN R + PV A +++
Sbjct 155 LKMSGNCLRASRPVLSFDDAFDKKPQLKL 183
> 7292473
Length=356
Score = 34.3 bits (77), Expect = 0.062, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 39/72 (54%), Gaps = 5/72 (6%)
Query 12 QTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKV--PLGPT--FIARLMNG 66
++K E+ + L + E+C+++ CN + E +RK D + ++ GP+ F+ ++
Sbjct 78 ESKMERSKTLSVVNEMCEMKHCNKAMLFEGRRKRDLYMWISNTSGSTGPSAKFLIENIHT 137
Query 67 LEVLRFSGNTER 78
+ L+ +GN R
Sbjct 138 MAELKMTGNCLR 149
> At3g15460
Length=315
Score = 33.5 bits (75), Expect = 0.11, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 36/62 (58%), Gaps = 3/62 (4%)
Query 17 KKEKLQSIAELCQLRSCNSVLYLEQRKN-DALLHVAKVPLGPT--FIARLMNGLEVLRFS 73
K + ++ EL +L+ +S L+ E RK+ D + + K P GP+ F+ ++ +E L+ +
Sbjct 90 KSSRGATLNELIELKGSSSCLFFECRKHKDLYMWMVKSPGGPSVKFLVNAVHTMEELKLT 149
Query 74 GN 75
GN
Sbjct 150 GN 151
> SPCC1259.09c
Length=456
Score = 32.0 bits (71), Expect = 0.26, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 1/54 (1%)
Query 43 KNDALLHVAKVPLG-PTFIARLMNGLEVLRFSGNTERTPPVGKLSAALSASANI 95
K D L HV K+ G P+ + + + LEVL FSG + P + S + S I
Sbjct 199 KGDVLAHVGKIDKGVPSSLQKFVRNLEVLDFSGVEPKKPSYTEDSVPVRKSTMI 252
> Hs8923081
Length=630
Score = 31.6 bits (70), Expect = 0.33, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 32/57 (56%), Gaps = 4/57 (7%)
Query 17 KKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAKVPLGPTFIARLMNGLEVLRFS 73
K+EK+Q + Q R+ N+ LY+E N+ + +P+ P F+ +L +G + + S
Sbjct 560 KEEKVQELLLRLQGRAENNCLYIEYGINEKI----TIPITPAFLGQLRSGRSIEKVS 612
> Hs22059448
Length=436
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 33/79 (41%), Gaps = 5/79 (6%)
Query 4 WALGFVLSQTKW-EKKEKLQSIAELC----QLRSCNSVLYLEQRKNDALLHVAKVPLGPT 58
W LGF Q + E++E+ + C Q C V L A ++V +PL P
Sbjct 191 WVLGFQTGQVTYCEEREEASGRSVPCLSAPQRELCGHVTQLRSAPVGAWVYVWSLPLRPL 250
Query 59 FIARLMNGLEVLRFSGNTE 77
G+ V+ G+TE
Sbjct 251 LYLCTTCGIMVITVKGDTE 269
> SPAC2G11.02
Length=1318
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Query 17 KKEKLQSIAELCQLRSCNSVLYLEQRKNDAL--LHVAKVPLGPTFIARLMNGLEVL 70
KKE L S C L CNSVL + +++L LH+ KV + I+ +N L L
Sbjct 916 KKESLCSGTFSCLLACCNSVLRTQNLTSESLNKLHILKVGIEEKAISLKLNELPFL 971
> At2g34810
Length=540
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 50 VAKVPLGPTFIARLMNGLEVLRFSGNTERTPPV 82
A +P +F LMNG+ LRF+ + R P V
Sbjct 53 TAVIPTNSSFSTNLMNGVRNLRFASVSTRKPEV 85
> Hs14971415
Length=1015
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Query 15 WEKKEKLQSIAELCQLRSCNSVLYLEQRKN-----DALLHVAKVPLGPTFIARLMNGLEV 69
+ KKE+L+ E C +C LE +++ + KV + T I +LM +
Sbjct 224 FHKKEQLKLYCETCDKLTCRDCQLLEHKEHRYQFIEEAFQNQKVIID-TLITKLMEKTKY 282
Query 70 LRFSGN 75
++F+GN
Sbjct 283 IKFTGN 288
> Hs17449320
Length=1050
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Query 15 WEKKEKLQSIAELCQLRSCNSVLYLEQRKN-----DALLHVAKVPLGPTFIARLMNGLEV 69
+ KKE+L+ E C +C LE +++ + KV + T I +LM +
Sbjct 225 FHKKEQLKLYCETCDKLTCRDCQLLEHKEHRYQFIEEAFQNQKVIID-TLITKLMEKTKY 283
Query 70 LRFSGN 75
++F+GN
Sbjct 284 IKFTGN 289
Lambda K H
0.322 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40