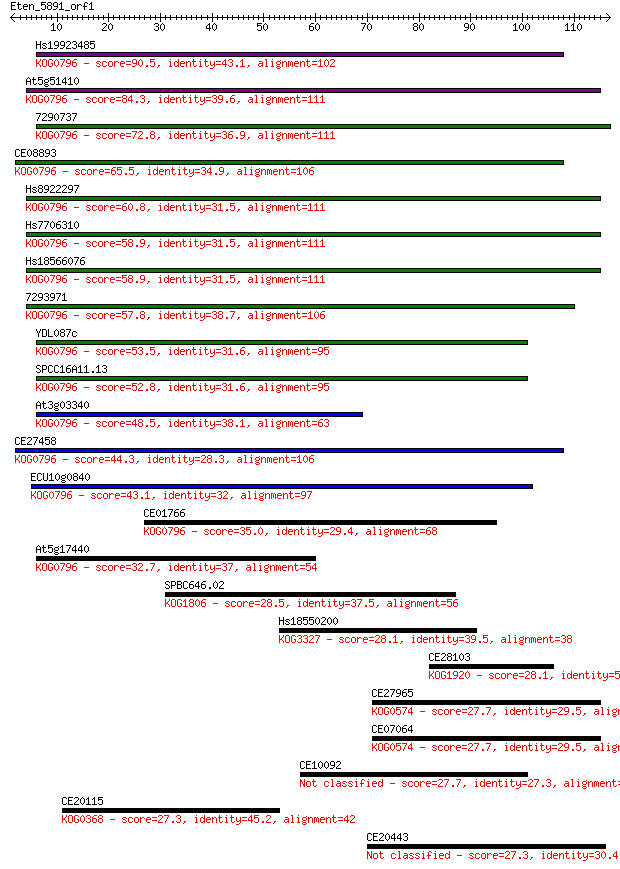
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5891_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
Hs19923485 90.5 6e-19
At5g51410 84.3 5e-17
7290737 72.8 1e-13
CE08893 65.5 2e-11
Hs8922297 60.8 6e-10
Hs7706310 58.9 2e-09
Hs18566076 58.9 2e-09
7293971 57.8 5e-09
YDL087c 53.5 1e-07
SPCC16A11.13 52.8 2e-07
At3g03340 48.5 3e-06
CE27458 44.3 6e-05
ECU10g0840 43.1 1e-04
CE01766 35.0 0.031
At5g17440 32.7 0.18
SPBC646.02 28.5 2.8
Hs18550200 28.1 3.7
CE28103 28.1 4.3
CE27965 27.7 5.0
CE07064 27.7 5.0
CE10092 27.7 5.4
CE20115 27.3 7.2
CE20443 27.3 7.5
> Hs19923485
Length=432
Score = 90.5 bits (223), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 44/102 (43%), Positives = 62/102 (60%), Gaps = 2/102 (1%)
Query 6 LDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRADLK 65
LD LMGRDRN + + +SVCK+YL FCP +LF NTR DLGPC++ H +L+
Sbjct 8 LDELMGRDRNLAPDEKRSNVRWDHESVCKYYLCGFCPAELFTNTRSDLGPCEKIHDENLR 67
Query 66 VAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRI 107
+E F + +E +F RYLQ L+ ++E RI+R R+
Sbjct 68 KQYEKSS--RFMKVGYERDFLRYLQSLLAEVERRIRRGHARL 107
> At5g51410
Length=334
Score = 84.3 bits (207), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 44/113 (38%), Positives = 64/113 (56%), Gaps = 2/113 (1%)
Query 4 ATLDSLMGRDRNEYGANAKKGASFKAD--SVCKFYLLDFCPLDLFPNTRVDLGPCDREHR 61
A LD LMG RN + K D VC FY++ FCP DLF NT+ DLG C R H
Sbjct 6 ALLDELMGAARNLTDEERRGFKEVKWDDREVCAFYMVRFCPHDLFVNTKSDLGACSRIHD 65
Query 62 ADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 114
LK +FE P + Y FE E A++ ++LV+ ++ +++R ++R+ +PV
Sbjct 66 PKLKESFENSPRHDSYVPKFEAELAQFCEKLVNDLDRKVRRGRERLAQEVEPV 118
> 7290737
Length=414
Score = 72.8 bits (177), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 41/116 (35%), Positives = 64/116 (55%), Gaps = 6/116 (5%)
Query 6 LDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRADLK 65
LD LMGR+RN + + A +++ C+FY + FCP DLF NTR DLGPC R H + +
Sbjct 9 LDELMGRNRNLHPSEAGAKVNWEDPEFCQFYNVKFCPHDLFINTRADLGPCARIHDEEAR 68
Query 66 VAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRI-----DANNQPVEL 116
+E D + +E EF R+ ++ ++ +I++ +QR+ D N P L
Sbjct 69 HLYE-DARPSQRKRQYEDEFLRFCNVMLHDVDRKIQKGKQRLLLMQRDQPNVPAPL 123
> CE08893
Length=369
Score = 65.5 bits (158), Expect = 2e-11, Method: Composition-based stats.
Identities = 37/107 (34%), Positives = 59/107 (55%), Gaps = 5/107 (4%)
Query 2 VCATLDSLMGRDRN-EYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREH 60
+ A LD LMG RN E G + K +F +C ++L+ FCP D+F NT+ DLG C H
Sbjct 7 MAAMLDELMGPKRNVELGKDTK--VTFDDPDICPYFLVGFCPHDMFINTKADLGACQLVH 64
Query 61 RADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRI 107
+L+ + P E+ Q FE R+L +L + + RI++ + ++
Sbjct 65 DDNLRRLYPESP--EYGQLGFERRLMRFLVQLDEDNQRRIRKNKDKL 109
> Hs8922297
Length=325
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 58/112 (51%), Gaps = 6/112 (5%)
Query 4 ATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRAD 63
A LD LMG R+ G ++ F D VCK +LLD CP D+ TR+DLG C + H
Sbjct 9 ALLDQLMGTARD--GDETRQRVKFTDDRVCKSHLLDCCPHDILAGTRMDLGECTKIHDLA 66
Query 64 LKVAFE-TDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 114
L+ +E ER+ + FE + +L+ + + + R + ++R+ + +
Sbjct 67 LRADYEIASKERDLF---FELDAMDHLESFIAECDRRTELAKKRLAETQEEI 115
> Hs7706310
Length=402
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 58/112 (51%), Gaps = 6/112 (5%)
Query 4 ATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRAD 63
A LD LMG R+ G ++ F D VCK +LL+ CP D+ TR+DLG C + H
Sbjct 9 AMLDQLMGTSRD--GDTTRQRIKFSDDRVCKSHLLNCCPHDVLSGTRMDLGECLKVHDLA 66
Query 64 LKVAFE-TDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 114
L+ +E E++F+ FE + +LQ + + R + ++R+ + +
Sbjct 67 LRADYEIASKEQDFF---FELDAMDHLQSFIADCDRRTEVAKKRLAETQEEI 115
> Hs18566076
Length=392
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 58/112 (51%), Gaps = 6/112 (5%)
Query 4 ATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRAD 63
A LD LMG R+ G ++ F D VCK +LL+ CP D+ TR+DLG C + H
Sbjct 9 AMLDQLMGTSRD--GDTTRQRIKFSDDRVCKSHLLNCCPHDVLSGTRMDLGECLKVHDLA 66
Query 64 LKVAFE-TDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 114
L+ +E E++F+ FE + +LQ + + R + ++R+ + +
Sbjct 67 LRADYEIASKEQDFF---FELDAMDHLQSFIADCDRRTEVAKKRLAETQEEI 115
> 7293971
Length=420
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 41/117 (35%), Positives = 59/117 (50%), Gaps = 21/117 (17%)
Query 4 ATLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREH--- 60
A LD LMG RN ++ F VCK +LLD CP D+ +TR+DLG C + H
Sbjct 16 AMLDQLMGTTRN----GDERQLKFSDPRVCKSFLLDCCPHDILASTRMDLGECPKVHDLA 71
Query 61 -RADLKVAFETDPEREFYQALFETEFARYLQ-------RLVDQMENRIKRVQQRIDA 109
RAD + A +T R++Y ++ E +LQ R D + R+K Q+ + A
Sbjct 72 FRADYESAAKT---RDYY---YDIEAMEHLQAFIADCDRRTDSAKQRLKETQEELTA 122
> YDL087c
Length=261
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 49/98 (50%), Gaps = 3/98 (3%)
Query 6 LDSLMGRD---RNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRA 62
++ LMGRD R+ ++ K+ +CK YL+ CP DLF T+ LG C + H
Sbjct 15 VEQLMGRDFSFRHNRYSHQKRDLGLHDPKICKSYLVGECPYDLFQGTKQSLGKCPQMHLT 74
Query 63 DLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRI 100
K+ +E + ++ FE E+ L R V++ +I
Sbjct 75 KHKIQYEREVKQGKTFPEFEREYLAILSRFVNECNGQI 112
> SPCC16A11.13
Length=264
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 48/95 (50%), Gaps = 4/95 (4%)
Query 6 LDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRADLK 65
++ LMG + + + ++ F VC+ +L CP D+F NT++DLGPC + H LK
Sbjct 9 IEQLMGSNLSNF--TSRGLVHFTDRKVCRSFLCGICPHDIFTNTKMDLGPCPKIHSDKLK 66
Query 66 VAFETDPEREFYQALFETEFARYLQRLVDQMENRI 100
+E Y +E ++ L+R VD RI
Sbjct 67 SDYERASYSHDYG--YEWDYLEDLERHVDDCNKRI 99
> At3g03340
Length=385
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 6 LDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRADLK 65
LD LMG +RN G + + VC+ YL CP DLF T++D+GPC + H L+
Sbjct 8 LDVLMGANRN--GDVQEVNRKYYDRDVCRLYLSGLCPHDLFQLTKMDMGPCPKVHSLQLR 65
Query 66 VAF 68
+
Sbjct 66 KEY 68
> CE27458
Length=339
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 58/109 (53%), Gaps = 8/109 (7%)
Query 2 VCATLDSLMGRDRNEYGANAKKGASFKAD--SVCKFYLLDFCPLDLFPNTRVDLGPCD-R 58
+ L+ LMG R+ AN + + D +VC +L+ FC D+F NT+ DLG C
Sbjct 5 MAQMLNELMGSQRD---ANPGERREIRYDDPNVCTDFLVGFCTHDIFRNTKNDLGFCKYT 61
Query 59 EHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRI 107
H +LK +++ ++ ++ FE F ++R+ + + +I++ + R+
Sbjct 62 THDENLKNSYKNSDKK--WRMGFEKRFLERIRRIHEDVRRKIQKHEDRL 108
> ECU10g0840
Length=245
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 49/97 (50%), Gaps = 4/97 (4%)
Query 5 TLDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDREHRADL 64
L+ L+G +R+ Y + + S K D VC + L+ FCP +LF NTR +G C +
Sbjct 8 VLNMLLGPERDTY--DPCRPTSTKKD-VCIYMLVSFCPFELFRNTRRSIGKCRYTSHEEY 64
Query 65 KVAFETDPEREFYQALFETEFARYLQRLVDQMENRIK 101
A RE + +E EF R L +V +++ I+
Sbjct 65 YKAEYNRNGRERAEE-YEWEFVRLLVEIVLSVQDGIR 100
> CE01766
Length=313
Score = 35.0 bits (79), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 27 FKADSVCKFYLLDFCPLDLFPNTRV-DLGPCDREHRADLKVAFE-TDPEREFYQALFETE 84
F SVC+ +LL CP D+ P++R+ ++ C + H K +E E++ + + E
Sbjct 28 FDHHSVCRAFLLGVCPHDMVPDSRLQNVVSCRKVHEPAHKADYERAQKEKDHFYDVDAFE 87
Query 85 FARYLQRLVD 94
+ LVD
Sbjct 88 IIEHAVHLVD 97
> At5g17440
Length=381
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 2/54 (3%)
Query 6 LDSLMGRDRNEYGANAKKGASFKADSVCKFYLLDFCPLDLFPNTRVDLGPCDRE 59
LD LMG +RN G + + VC+ YL CP +LF T + DRE
Sbjct 8 LDVLMGANRN--GDVTEVNRKYYDRDVCRLYLSGLCPHELFQLTAKGVDNYDRE 59
> SPBC646.02
Length=1284
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 28/63 (44%), Gaps = 7/63 (11%)
Query 31 SVCK-FYLLDFCPLDLFPNTRVDLGPCDREHRAD------LKVAFETDPEREFYQALFET 83
S+C Y LD +D PN R+D G H K + ET+P + Q L E
Sbjct 1054 SLCSSIYPLDIKTVDSSPNKRLDYGNSGFAHEVQFINVGAFKGSQETEPVSGYKQNLGEA 1113
Query 84 EFA 86
E+A
Sbjct 1114 EYA 1116
> Hs18550200
Length=449
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 19/38 (50%), Gaps = 1/38 (2%)
Query 53 LGPCDREHRADLKVAFETDPEREFYQALFETEFARYLQ 90
LG C R L FE DP + +Q L+E + R LQ
Sbjct 150 LGACQEAPRPHLG-EFEADPRGQLWQRLWEVQDGRRLQ 186
> CE28103
Length=1250
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 82 ETEFARYLQRLVDQMENRIKRVQQ 105
+TEF Y +RL EN++KRV+Q
Sbjct 1063 KTEFENYKKRLAVVRENKLKRVEQ 1086
> CE27965
Length=525
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 71 DPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 114
D + EF + + E R + L +ME I+ +Q+R QP+
Sbjct 471 DGDFEFLRNITLDELIRRKESLDSEMEEEIRELQRRYKTKRQPI 514
> CE07064
Length=523
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 71 DPEREFYQALFETEFARYLQRLVDQMENRIKRVQQRIDANNQPV 114
D + EF + + E R + L +ME I+ +Q+R QP+
Sbjct 469 DGDFEFLRNITLDELIRRKESLDSEMEEEIRELQRRYKTKRQPI 512
> CE10092
Length=5105
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 12/44 (27%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 57 DREHRADLKVAFETDPEREFYQALFETEFARYLQRLVDQMENRI 100
DRE R + + + E RE +A+FE + Q ++++ E +
Sbjct 1384 DRESRYNEEQSLEMKNRRELQRAIFEKRIITFRQPVIEETEQEL 1427
> CE20115
Length=1657
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 24/49 (48%), Gaps = 10/49 (20%)
Query 11 GRDRNEYGANAKKG-------ASFKADSVCKFYLLDFCPLDLFPNTRVD 52
GRDRNE AN K G A+F S YL+D L+ F + + D
Sbjct 461 GRDRNEAIANLKHGLQNLKIDATFPTQSD---YLIDLLSLEKFKSNQYD 506
> CE20443
Length=727
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 26/53 (49%), Gaps = 7/53 (13%)
Query 70 TDPEREFYQALF-------ETEFARYLQRLVDQMENRIKRVQQRIDANNQPVE 115
T+P ++F+Q F E ++ Y + LV + +I R+ I N + V+
Sbjct 98 TEPSKQFFQEQFVKEWQYHEIDYTPYFETLVPRFREKIARLLDEIVKNPEEVD 150
Lambda K H
0.323 0.138 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40