bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5940_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
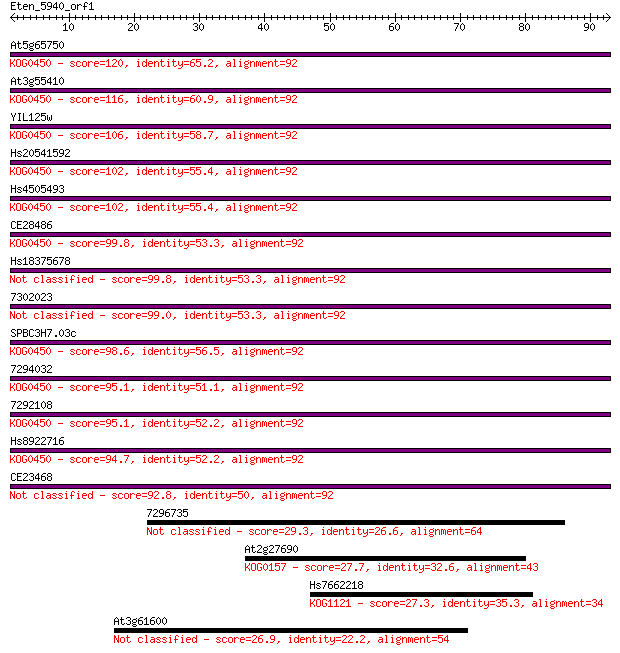
At5g65750 120 7e-28
At3g55410 116 1e-26
YIL125w 106 1e-23
Hs20541592 102 1e-22
Hs4505493 102 2e-22
CE28486 99.8 1e-21
Hs18375678 99.8 1e-21
7302023 99.0 2e-21
SPBC3H7.03c 98.6 3e-21
7294032 95.1 3e-20
7292108 95.1 3e-20
Hs8922716 94.7 4e-20
CE23468 92.8 1e-19
7296735 29.3 2.0
At2g27690 27.7 5.3
Hs7662218 27.3 7.6
At3g61600 26.9 10.0
> At5g65750
Length=1025
Score = 120 bits (300), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 60/92 (65%), Positives = 70/92 (76%), Gaps = 2/92 (2%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W RQ GLV+LLPHGYDGQGPEHSSGR+ERFLQ+ DD VI E + L + IQ+ N Q
Sbjct 758 WLRQTGLVVLLPHGYDGQGPEHSSGRLERFLQMSDDNPYVIPEMDPTLRKQ--IQECNWQ 815
Query 61 VVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
VV T P+N FHVLRRQ+HR FRKPL+V +PK
Sbjct 816 VVNVTTPANYFHVLRRQIHRDFRKPLIVMAPK 847
> At3g55410
Length=1009
Score = 116 bits (290), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 56/92 (60%), Positives = 68/92 (73%), Gaps = 2/92 (2%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W RQ GLV+LLPHGYDGQGPEHSS R+ER+LQ+ DD VI + E + IQ+ N Q
Sbjct 754 WLRQTGLVMLLPHGYDGQGPEHSSARLERYLQMSDDNPYVI--PDMEPTMRKQIQECNWQ 811
Query 61 VVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
+V T P+N FHVLRRQ+HR FRKPL+V +PK
Sbjct 812 IVNATTPANYFHVLRRQIHRDFRKPLIVMAPK 843
> YIL125w
Length=1014
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 54/92 (58%), Positives = 64/92 (69%), Gaps = 3/92 (3%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W +++GLVL LPHGYDGQGPEHSSGR+ERFLQL ++ E +L R Q N Q
Sbjct 763 WKQRSGLVLSLPHGYDGQGPEHSSGRLERFLQLANEDPRYFPSEE-KLQRQH--QDCNFQ 819
Query 61 VVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
VV PT P+N FH+LRRQ HR FRKPL +F K
Sbjct 820 VVYPTTPANLFHILRRQQHRQFRKPLALFFSK 851
> Hs20541592
Length=1023
Score = 102 bits (255), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 51/95 (53%), Positives = 67/95 (70%), Gaps = 6/95 (6%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDV---IHEENWELSRSSVIQQH 57
W RQNG+VLLLPHG +G GPEHSS R ERFLQ+C+D DV + E N+++++ +
Sbjct 769 WVRQNGIVLLLPHGMEGMGPEHSSARPERFLQMCNDDPDVLPDLKEANFDINQ---LYDC 825
Query 58 NIQVVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
N VV + P N FHVLRRQ+ FRKPL++F+PK
Sbjct 826 NWVVVNCSTPGNFFHVLRRQILLPFRKPLIIFTPK 860
> Hs4505493
Length=1002
Score = 102 bits (254), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/95 (53%), Positives = 67/95 (70%), Gaps = 6/95 (6%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDV---IHEENWELSRSSVIQQH 57
W RQNG+VLLLPHG +G GPEHSS R ERFLQ+C+D DV + E N+++++ +
Sbjct 769 WVRQNGIVLLLPHGMEGMGPEHSSARPERFLQMCNDDPDVLPDLKEANFDINQ---LYDC 825
Query 58 NIQVVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
N VV + P N FHVLRRQ+ FRKPL++F+PK
Sbjct 826 NWVVVNCSTPGNFFHVLRRQILLPFRKPLIIFTPK 860
> CE28486
Length=1029
Score = 99.8 bits (247), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 49/94 (52%), Positives = 64/94 (68%), Gaps = 2/94 (2%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W RQ+GLV+LLPHGY+G GPEHSS R ERFLQ+C++ +++ E+ Q H+
Sbjct 769 WIRQSGLVMLLPHGYEGMGPEHSSARPERFLQMCNEDDEIDLEKIAFEGTFEAQQLHDTN 828
Query 61 VVVP--TVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
+V T P+N +H+LRRQV FRKP VVFSPK
Sbjct 829 WIVANCTTPANIYHLLRRQVTMPFRKPAVVFSPK 862
> Hs18375678
Length=919
Score = 99.8 bits (247), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 49/92 (53%), Positives = 62/92 (67%), Gaps = 11/92 (11%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W Q+G+V+LLPHGYDG GP+HSS RIERFLQ+CD E+ + + N+
Sbjct 687 WLLQSGIVILLPHGYDGAGPDHSSCRIERFLQMCDSAEEGVDGDTV-----------NMF 735
Query 61 VVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
VV PT P+ FH+LRRQ+ R FRKPL+V SPK
Sbjct 736 VVHPTTPAQYFHLLRRQMVRNFRKPLIVASPK 767
> 7302023
Length=901
Score = 99.0 bits (245), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 49/92 (53%), Positives = 58/92 (63%), Gaps = 11/92 (11%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W N LV+LLPHGYDG EHSS RIERFLQLCD +E S+ N+
Sbjct 670 WMESNALVMLLPHGYDGAASEHSSCRIERFLQLCDSKET-----------SADGDSVNVH 718
Query 61 VVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
+V PT P+ +HVLRRQ+ R FRKPLVV +PK
Sbjct 719 IVNPTTPAQYYHVLRRQLARNFRKPLVVVAPK 750
> SPBC3H7.03c
Length=1009
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 52/93 (55%), Positives = 63/93 (67%), Gaps = 5/93 (5%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLC-DDREDVIHEENWELSRSSVIQQHNI 59
W ++ G+VL LPHGYDGQGPEHSS R+ER+LQLC +D + EE +L R Q NI
Sbjct 761 WLQRTGIVLSLPHGYDGQGPEHSSARMERYLQLCNEDPREFPSEE--KLQRQH--QDCNI 816
Query 60 QVVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
Q + T PS FH LRR +HR FRKPLV+F K
Sbjct 817 QAIYVTKPSQYFHALRRNIHRQFRKPLVIFFSK 849
> 7294032
Length=1001
Score = 95.1 bits (235), Expect = 3e-20, Method: Composition-based stats.
Identities = 47/94 (50%), Positives = 63/94 (67%), Gaps = 5/94 (5%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W RQ+GLV+LLPHG +G GPEHSS R+ERFLQ+ D D E+ E V Q H+I
Sbjct 762 WVRQSGLVMLLPHGMEGMGPEHSSCRVERFLQMSSDDPDYFPPESDEF---GVRQLHDIN 818
Query 61 VVVP--TVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
+V + P+N +H+LRRQ+ FRKPL++ +PK
Sbjct 819 WIVANCSTPANYYHILRRQIALPFRKPLILCTPK 852
> 7292108
Length=1075
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 59/92 (64%), Gaps = 0/92 (0%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W RQ+G+V+LLPH +G GPEHSSGRIERFLQ+ DD DV + + + N
Sbjct 785 WVRQSGVVMLLPHSMEGMGPEHSSGRIERFLQMSDDDPDVYPDTCDADFVARQLMNVNWI 844
Query 61 VVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
V + P+N FH LRRQV FRKPL+ FSPK
Sbjct 845 VTNLSTPANLFHCLRRQVKMGFRKPLINFSPK 876
> Hs8922716
Length=1010
Score = 94.7 bits (234), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 48/94 (51%), Positives = 64/94 (68%), Gaps = 5/94 (5%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDV--IHEENWELSRSSVIQQHN 58
W R NG+VLLLPHG +G GPEHSS R ERFLQ+ +D D +++E+S+ + N
Sbjct 756 WVRHNGIVLLLPHGMEGMGPEHSSARPERFLQMSNDDSDAYPAFTKDFEVSQ---LYDCN 812
Query 59 IQVVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
VV + P+N FHVLRRQ+ FRKPL++F+PK
Sbjct 813 WIVVNCSTPANYFHVLRRQILLPFRKPLIIFTPK 846
> CE23468
Length=906
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/92 (50%), Positives = 58/92 (63%), Gaps = 9/92 (9%)
Query 1 WNRQNGLVLLLPHGYDGQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQ 60
W +GL +LLPHG+DG GPEHSS R+ERFLQLCD RED + + N++
Sbjct 672 WLTSSGLTMLLPHGFDGAGPEHSSCRMERFLQLCDSREDQTPVDG---------ENVNMR 722
Query 61 VVVPTVPSNTFHVLRRQVHRAFRKPLVVFSPK 92
V PT + FH+LRRQV +RKPL+V PK
Sbjct 723 VANPTTSAQYFHLLRRQVVPNYRKPLIVVGPK 754
> 7296735
Length=1247
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 33/64 (51%), Gaps = 1/64 (1%)
Query 22 HSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQVVVPTVPSNTFHVLRRQVHRA 81
H+S + R+ ++ + DVI + +L + + N+QV +P + F+ R ++
Sbjct 965 HASLDLMRYTEMVKNANDVIGSKGLDLLGNVYAPRGNVQVHLPFLEVTAFNP-RSRITLT 1023
Query 82 FRKP 85
FRKP
Sbjct 1024 FRKP 1027
> At2g27690
Length=495
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 24/48 (50%), Gaps = 5/48 (10%)
Query 37 REDVIHEENW-----ELSRSSVIQQHNIQVVVPTVPSNTFHVLRRQVH 79
++D I+ +W S +S I+ H + V+ PSN H+L+ H
Sbjct 46 KKDFINLSDWYTHLLRRSPTSTIKVHVLNSVITANPSNVEHILKTNFH 93
> Hs7662218
Length=1171
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 47 ELSRSSVIQQHNIQVVVPTVPSNTFHVLRRQVHR 80
EL R + QH++ VP+ S +FH+L R + +
Sbjct 872 ELQREYALPQHHLIQDVPSKWSTSFHMLERLIEQ 905
> At3g61600
Length=545
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 12/54 (22%), Positives = 24/54 (44%), Gaps = 0/54 (0%)
Query 17 GQGPEHSSGRIERFLQLCDDREDVIHEENWELSRSSVIQQHNIQVVVPTVPSNT 70
G+ P+ G L D E+ E NW + S+V++ + + P + + +
Sbjct 98 GENPDDEGGEAMVEEALSGDEEETSSEPNWGMDCSTVVRVKELHISSPILAAKS 151
Lambda K H
0.322 0.138 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171209254
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40