bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6007_orf2
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
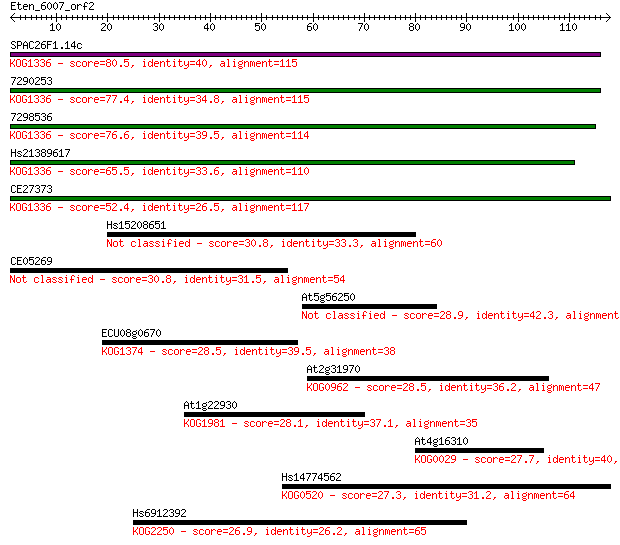
SPAC26F1.14c 80.5 7e-16
7290253 77.4 6e-15
7298536 76.6 9e-15
Hs21389617 65.5 2e-11
CE27373 52.4 2e-07
Hs15208651 30.8 0.58
CE05269 30.8 0.62
At5g56250 28.9 2.4
ECU08g0670 28.5 3.2
At2g31970 28.5 3.3
At1g22930 28.1 4.2
At4g16310 27.7 5.9
Hs14774562 27.3 6.7
Hs6912392 26.9 8.0
> SPAC26F1.14c
Length=256
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 46/130 (35%), Positives = 70/130 (53%), Gaps = 15/130 (11%)
Query 1 HYGAPLAKGVATAD-HVTCPWHDAKFDLKTGKCIAGPVTQGIQTYPVQVKGS-SLFASLP 58
HYGAPLAKGV T+D H+ CPWH A F+ TG P ++T+PV +G SL+ +
Sbjct 63 HYGAPLAKGVVTSDGHIVCPWHGACFNAATGDVEDTPAIAALRTFPVTEEGDGSLWIEVE 122
Query 59 ERLEEHEHSIS---CCKRRETAFKDKTFV----------IVGGGPAASAAVEELRAQGFE 105
++ + + C + + T +K V I+GGG AS A E LR + F+
Sbjct 123 DKNDNGASVLQPEGCWRNKATEVYNKGSVETEVTAPHVCIIGGGKGASVAAEYLREKNFK 182
Query 106 GRLVLISREN 115
G++ + +RE+
Sbjct 183 GKITIFTRED 192
> 7290253
Length=552
Score = 77.4 bits (189), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 62/117 (52%), Gaps = 4/117 (3%)
Query 1 HYGAPLAKGVATADHVTCPWHDAKFDLKTGKCIAGPVTQGIQTYPVQV--KGSSLFASLP 58
HYGAPL G V CPWH A F+L+ G P + Y V+V +G + +
Sbjct 59 HYGAPLQTGALGLGRVRCPWHGACFNLENGDIEDFPGLDSLPCYRVEVGNEGQVMLRAKR 118
Query 59 ERLEEHEHSISCCKRRETAFKDKTFVIVGGGPAASAAVEELRAQGFEGRLVLISREN 115
L ++ + +R+ + F++VGGGP+ + AVE +R +GF GRL+ + RE+
Sbjct 119 SDLVNNKRLKNMVRRKPD--DQRVFIVVGGGPSGAVAVETIRQEGFTGRLIFVCRED 173
> 7298536
Length=539
Score = 76.6 bits (187), Expect = 9e-15, Method: Composition-based stats.
Identities = 45/118 (38%), Positives = 62/118 (52%), Gaps = 8/118 (6%)
Query 1 HYGAPLAKGVATADHVTCPWHDAKFDLKTGKCIAGPVTQGIQTYPVQV--KGSSLFASLP 58
H GAPLAKGV + + V CPWH F+L+TG P + + V V +G L
Sbjct 52 HRGAPLAKGVLSRNRVRCPWHGVCFNLETGDIENFPGLDSLPCHRVNVDSRGQVLVQVKR 111
Query 59 ERLEEHEH--SISCCKRRETAFKDKTFVIVGGGPAASAAVEELRAQGFEGRLVLISRE 114
L H S++C ++ + FV+VGGGP+ + VE LR +GF GRL L+ E
Sbjct 112 SYLLRHARIKSMACRNWQD----QRHFVVVGGGPSGAVCVETLRQEGFTGRLTLVCGE 165
> Hs21389617
Length=605
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 55/114 (48%), Gaps = 4/114 (3%)
Query 1 HYGAPLAKGVATADHVTCPWHDAKFDLKTGKCIAGPVTQGIQTYPVQVKGSSLFASLPE- 59
HYGAPL KGV + V CPWH A F++ TG P + + V+++ ++ +
Sbjct 111 HYGAPLVKGVLSRGRVRCPWHGACFNISTGDLEDFPGLDSLHKFQVKIEKEKVYVRASKQ 170
Query 60 --RLEEHEHSISCCKRRETAFKDKTFV-IVGGGPAASAAVEELRAQGFEGRLVL 110
+L+ ++ C + T V IVG G A E LR +GF R+VL
Sbjct 171 ALQLQRRTKVMAKCISPSAGYSSSTNVLIVGAGAAGLVCAETLRQEGFSDRIVL 224
> CE27373
Length=549
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 52/117 (44%), Gaps = 1/117 (0%)
Query 1 HYGAPLAKGVATADHVTCPWHDAKFDLKTGKCIAGPVTQGIQTYPVQVKGSSLFASLPER 60
HY L G + CP H A F++++G P + +Y V V +L E+
Sbjct 65 HYNFSLENGTYAKGRIRCPLHGACFNVRSGDIEDYPGFDSLHSYVVTVNDGNLIIKTTEK 124
Query 61 LEEHEHSISCCKRRETAFKDKTFVIVGGGPAASAAVEELRAQGFEGRLVLISRENVP 117
+ I + + D+ VI+GGG A + +E R G +++IS E++P
Sbjct 125 KLGSDRRIRHLPKMKQC-NDRPVVIIGGGVATATFIEHSRLNGLITPILVISEESLP 180
> Hs15208651
Length=417
Score = 30.8 bits (68), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 28/62 (45%), Gaps = 3/62 (4%)
Query 20 WHDAKFDLKTGKCIAGPVTQGIQTYPVQVKGSSLFAS--LPERLEEHEHSISCCKRRETA 77
WH + TG+C A + + Q + + S+LF S LP RL H R E A
Sbjct 209 WHSMASNY-TGRCHAATLARNTQLHWLWAASSALFPSIYLPPRLPPAHHQAFVRHRLEEA 267
Query 78 FK 79
F+
Sbjct 268 FR 269
> CE05269
Length=474
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 30/63 (47%), Gaps = 9/63 (14%)
Query 1 HYGAPLAKG--VATADHVTCPWHDAKFDLKTGKCIAGPVTQG-------IQTYPVQVKGS 51
H GA G V + + CP+H F +TGKC+ P +G + T+P + +
Sbjct 170 HIGANFNIGGRVVRDNCIQCPFHGWIFSAETGKCVEVPYDEGRIPEQAKVTTWPCIERNN 229
Query 52 SLF 54
+++
Sbjct 230 NIY 232
> At5g56250
Length=811
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 17/26 (65%), Gaps = 1/26 (3%)
Query 58 PERLEEHEHSISCCKRRETAFKDKTF 83
P R +++ S CC+R+E A +D TF
Sbjct 691 PSRFRDNDQS-CCCQRKEKALEDTTF 715
> ECU08g0670
Length=434
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 19/38 (50%), Gaps = 6/38 (15%)
Query 19 PWHDAKFDLKTGKCIAGPVTQGIQTYPVQVKGSSLFAS 56
PW F++ GKCIA +T P +V G SL S
Sbjct 341 PWMPPSFNVALGKCIAN------ETRPSRVSGLSLTNS 372
> At2g31970
Length=1292
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 59 ERLEEHEHSISCCKRRETAFKDKTFVIVGGGPAASAAVEELRAQGFE 105
E+ EHS CC+R TA ++ +F I AS+ E L+A E
Sbjct 670 EKRARQEHSCPCCERSFTADEEASF-IKKQRVKASSTGEHLKALAVE 715
> At1g22930
Length=1131
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 35 GPVTQGIQTYPVQVKGSSLFASLPERLEEHEHSIS 69
GP TQ ++ PV + S S E EEH +++S
Sbjct 859 GPPTQAYESLPVTRRWISTLLSSKEEWEEHNNTLS 893
> At4g16310
Length=1265
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 80 DKTFVIVGGGPAASAAVEELRAQGF 104
+K +++G GPA A L+ QGF
Sbjct 617 EKKVIVIGAGPAGLTAARHLQRQGF 641
> Hs14774562
Length=1202
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 33/75 (44%), Gaps = 11/75 (14%)
Query 54 FASLPE------RLEEHEHSISCCKRRETAFKDKTFVIVGG-----GPAASAAVEELRAQ 102
F SLP L++++ +S +R E K + G GP A +E +
Sbjct 619 FLSLPSTQLDWLSLDDNQFRMSILERLEQMEKRMAEIAAAGQVPCQGPDAPPVQDEGQGP 678
Query 103 GFEGRLVLISRENVP 117
GFE R+V++ +P
Sbjct 679 GFEARVVVLVESMIP 693
> Hs6912392
Length=558
Score = 26.9 bits (58), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 30/66 (45%), Gaps = 3/66 (4%)
Query 25 FDLKTGKCIAG-PVTQGIQTYPVQVKGSSLFASLPERLEEHEHSISCCKRRETAFKDKTF 83
+D+ C+ G P++QG + G +F + + ++ S F+DKTF
Sbjct 247 YDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFI--NQASYMSILGMTPGFRDKTF 304
Query 84 VIVGGG 89
V+ G G
Sbjct 305 VVQGFG 310
Lambda K H
0.318 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40