bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6025_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
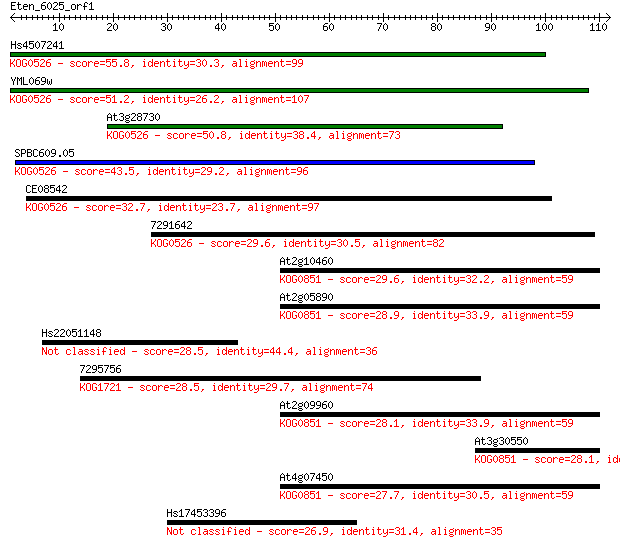
Hs4507241 55.8 2e-08
YML069w 51.2 4e-07
At3g28730 50.8 6e-07
SPBC609.05 43.5 1e-04
CE08542 32.7 0.15
7291642 29.6 1.3
At2g10460 29.6 1.3
At2g05890 28.9 2.3
Hs22051148 28.5 2.8
7295756 28.5 3.6
At2g09960 28.1 4.2
At3g30550 28.1 4.6
At4g07450 27.7 5.0
Hs17453396 26.9 9.1
> Hs4507241
Length=709
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 4/99 (4%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
+E + F H++L+L K++ +WG V G L+ + P FE+ +++Q TT
Sbjct 82 FEKLSDFFKTHYRLELMEKDLCVKGWNWGTVKFGGQLLSFDIGDQPVFEIPLSNVSQCTT 141
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETD 99
KN++ +E Q+D + S L+E+R + P T +D D
Sbjct 142 -GKNEVTLEFHQNDDAEVS---LMEVRFYVPPTQEDGVD 176
> YML069w
Length=552
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 1/108 (0%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
Y I+ F F +Q++ + + +WG + N + ++NG P FE+ I
Sbjct 86 YNLIKNDFHRRFNIQVEQREHSLRGWNWGKTDLARNEMVFALNGKPTFEIPYARINNTNL 145
Query 61 PSKNDLAIEL-VQDDTMDQSEDMLLEIRLFQPATGDDETDSHLQNLKE 107
SKN++ IE +QD+ + D L+E+R + P D ++ +E
Sbjct 146 TSKNEVGIEFNIQDEEYQPAGDELVEMRFYIPGVIQTNVDENMTKKEE 193
> At3g28730
Length=646
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 40/74 (54%), Gaps = 1/74 (1%)
Query 19 KNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPSKNDLAIELVQDDTMDQ 78
K ++ R+WG+V + GN L V AFEV+ D++Q KND+ +E DDT
Sbjct 100 KQLSVSGRNWGEVDLHGNTLTFLVGSKQAFEVSLADVSQTQLQGKNDVTLEFHVDDTAGA 159
Query 79 SE-DMLLEIRLFQP 91
+E D L+EI P
Sbjct 160 NEKDSLMEISFHIP 173
> SPBC609.05
Length=512
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 28/99 (28%), Positives = 47/99 (47%), Gaps = 3/99 (3%)
Query 2 EDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTP 61
+D+ +F + ++ K + +WG+ + G+ L VN PAFE+ +
Sbjct 82 DDLINVIKQNFDMGIEQKEFSIKGWNWGEANFLGSELVFDVNSRPAFEIPISAVTNTNLS 141
Query 62 SKNDLAIEL-VQDDTMDQSE--DMLLEIRLFQPATGDDE 97
KN++A+E DD S D L+E+RL+ P T E
Sbjct 142 GKNEVALEFSTTDDKQIPSAQVDELVEMRLYVPGTTAKE 180
> CE08542
Length=689
Score = 32.7 bits (73), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 23/97 (23%), Positives = 47/97 (48%), Gaps = 4/97 (4%)
Query 4 IRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPSK 63
I+ S ++ + N+ ++G + G N+ S P FE+ +++ V +K
Sbjct 84 IQSFTSSNWSKSINQSNLFINGWNYGQADVKGKNIEFSWENEPIFEIPCTNVSNVIA-NK 142
Query 64 NDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETDS 100
N+ +E Q+ +QS+ L+E+R P ++E D+
Sbjct 143 NEAILEFHQN---EQSKVQLMEMRFHMPVDLENEEDT 176
> 7291642
Length=723
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 37/85 (43%), Gaps = 7/85 (8%)
Query 27 HWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPSKNDLAIELVQDDTMDQSEDMLLEI 86
+WG G+ L+ FEV ++Q T KN++ +E Q+D LLE+
Sbjct 108 NWGTARFMGSVLSFDKESKTIFEVPLSHVSQCVT-GKNEVTLEFHQNDDAPVG---LLEM 163
Query 87 RLFQPA---TGDDETDSHLQNLKEK 108
R PA +D D QN+ K
Sbjct 164 RFHIPAVESAEEDPVDKFHQNVMSK 188
> At2g10460
Length=251
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 31/62 (50%), Gaps = 3/62 (4%)
Query 51 NAQDIAQVTT-PSKNDLAI--ELVQDDTMDQSEDMLLEIRLFQPATGDDETDSHLQNLKE 107
N+ D++QV P+ +L + + + D + RL+ P TGDD+ D Q +KE
Sbjct 92 NSFDVSQVFVDPTLAELGLFKQSIPTDGLTLGSSGSFHKRLYAPKTGDDDGDYPRQTIKE 151
Query 108 KL 109
L
Sbjct 152 VL 153
> At2g05890
Length=456
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 3/62 (4%)
Query 51 NAQDIAQV---TTPSKNDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETDSHLQNLKE 107
N D++QV T ++ DL + + D + RL+ P TGDD+ D Q +KE
Sbjct 192 NFFDVSQVFVDPTLAELDLFKQSIPTDGLTLGSSGSFHKRLYAPRTGDDDGDYPRQTIKE 251
Query 108 KL 109
L
Sbjct 252 VL 253
> Hs22051148
Length=1648
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 7 HFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSV 42
H S+H KLQ +T N HL + S+ GNNL S+
Sbjct 51 HHSEH-KLQTKTLNTLHLQSEPPECSIGGNNLENSL 85
> 7295756
Length=714
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 40/74 (54%), Gaps = 6/74 (8%)
Query 14 LQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPSKNDLAIELVQD 73
+QL +N ++S H DV+ G +N+ ++G ++NA + +TP+ ++ +V D
Sbjct 238 VQLNFENFHNISHHVNDVANVG--VNMVLHGQATDKLNAVVVNTSSTPN----SVPVVDD 291
Query 74 DTMDQSEDMLLEIR 87
+EDML IR
Sbjct 292 VEFSDTEDMLEGIR 305
> At2g09960
Length=345
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 3/62 (4%)
Query 51 NAQDIAQVTTPSKND---LAIELVQDDTMDQSEDMLLEIRLFQPATGDDETDSHLQNLKE 107
N+ D++QV S D L + + D + RL+ P TGDD+ D Q +KE
Sbjct 132 NSFDVSQVFVDSTLDELGLFKQSIPIDGLTLESSGSFHKRLYAPRTGDDDGDYPRQIIKE 191
Query 108 KL 109
L
Sbjct 192 VL 193
> At3g30550
Length=509
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 87 RLFQPATGDDETDSHLQNLKEKL 109
RL+ P TGDD+ D Q +KE L
Sbjct 251 RLYAPRTGDDDGDYPRQTIKEVL 273
> At4g07450
Length=439
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 31/62 (50%), Gaps = 3/62 (4%)
Query 51 NAQDIAQVTT-PSKNDLAI--ELVQDDTMDQSEDMLLEIRLFQPATGDDETDSHLQNLKE 107
N+ D++QV P+ +L + + + D + RL+ P TGDD+ D Q +K+
Sbjct 141 NSFDVSQVFVDPTLAELGLFKQSIPTDGLTLGSSGSFHKRLYAPRTGDDDGDYPRQTIKD 200
Query 108 KL 109
L
Sbjct 201 VL 202
> Hs17453396
Length=200
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 11/35 (31%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 30 DVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPSKN 64
+S D + V+++ CPAF + +TTP K+
Sbjct 151 SLSQDETCIKVAMSKCPAFPGEPSTVPDLTTPRKS 185
Lambda K H
0.315 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40