bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6086_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
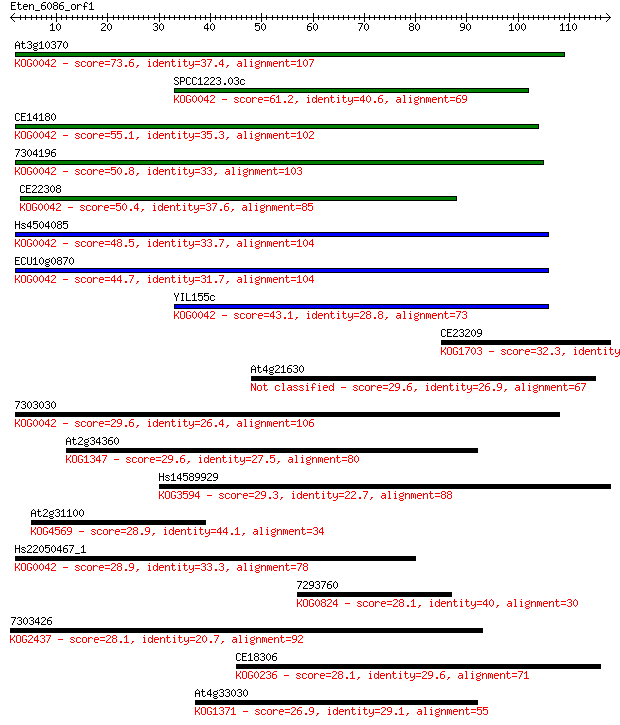
At3g10370 73.6 8e-14
SPCC1223.03c 61.2 5e-10
CE14180 55.1 3e-08
7304196 50.8 6e-07
CE22308 50.4 8e-07
Hs4504085 48.5 3e-06
ECU10g0870 44.7 4e-05
YIL155c 43.1 1e-04
CE23209 32.3 0.20
At4g21630 29.6 1.3
7303030 29.6 1.5
At2g34360 29.6 1.5
Hs14589929 29.3 2.1
At2g31100 28.9 2.3
Hs22050467_1 28.9 2.5
7293760 28.1 4.3
7303426 28.1 4.4
CE18306 28.1 4.5
At4g33030 26.9 8.2
> At3g10370
Length=629
Score = 73.6 bits (179), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 60/107 (56%), Gaps = 0/107 (0%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPI 61
+HL YG MA V +A++E L + L G P+L+AEV Y R+E S D +ARR I
Sbjct 512 KHLSHAYGSMADRVATIAQEEGLGKRLAHGHPFLEAEVAYCARHEYCESAVDFIARRCRI 571
Query 62 LFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFTAA 108
FLD A+ V ++A E W + ++ Q+AK++ ETF ++
Sbjct 572 AFLDTDAAARALQRVVEILASEHKWDKSRQKQELQKAKEFLETFKSS 618
> SPCC1223.03c
Length=649
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 43/69 (62%), Gaps = 0/69 (0%)
Query 33 PYLKAEVLYACRYEQARSVSDILARRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATAN 92
P+ AE+ Y+ +YE R+ +D LARR + FLD + ++A+ V +M +E GW AT +
Sbjct 562 PFTVAELKYSIKYEYTRTPTDFLARRTRLAFLDARQALQAVAGVTHVMKEEFGWDDATTD 621
Query 93 KKKQEAKKY 101
K QEA+ Y
Sbjct 622 KLAQEARDY 630
> CE14180
Length=722
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 55/107 (51%), Gaps = 6/107 (5%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEPLVEG-----LPYLKAEVLYACRYEQARSVSDILA 56
+HL YG A V + K P+V PYL AEV YA R E A + D++A
Sbjct 497 QHLSNTYGDRAFVVARMCKMTGKRWPIVGQRLHPEFPYLDAEVRYAVR-EYACTAIDVIA 555
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAE 103
RR+ + FL+ E + V +M +ELGWS+A + ++A+ + +
Sbjct 556 RRMRLAFLNTYAAHEVLPDVVRVMGQELGWSSAEQRAQLEKARTFID 602
> 7304196
Length=713
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 55/108 (50%), Gaps = 6/108 (5%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEPLV-----EGLPYLKAEVLYACRYEQARSVSDILA 56
EHL YG A V A P+V PY++AEV R + A ++ D++A
Sbjct 477 EHLSNTYGSNAFKVAVSADSSGKAWPIVGKRLHSEFPYIEAEVKQGVR-DFACTLEDMVA 535
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAET 104
RRL + FL+ ++ E + V ++MAKEL WS K+ ++ + + +
Sbjct 536 RRLRVAFLNVQVAEEILPQVANIMAKELNWSRDQIKKQIRQTRTFLNS 583
> CE22308
Length=673
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 50/90 (55%), Gaps = 6/90 (6%)
Query 3 HLVRHYGFMARDVVELAKKESLMEPLVE-----GLPYLKAEVLYACRYEQARSVSDILAR 57
HL + YG +V++L K P++ P+L+AEV YA + E AR +DILAR
Sbjct 486 HLCQTYGDKVYEVLKLCKSTGQKFPVIGHRLHPDFPFLEAEVRYAVK-EYARIPADILAR 544
Query 58 RLPILFLDHKLGVEAIDTVCSMMAKELGWS 87
R + LD + + + + ++MA+EL WS
Sbjct 545 RTRLSLLDARAARQVLPRIVAIMAEELKWS 574
> Hs4504085
Length=727
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 55/109 (50%), Gaps = 10/109 (9%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEP-----LVEGLPYLKAEVLYACRYEQARSVSDILA 56
+HL YG A +V ++A P LV PY++AEV Y + E A + D+++
Sbjct 495 QHLAATYGDKAFEVAKMASVTGKRWPIVGVHLVSEFPYIEAEVKYGIK-EYACTAVDMIS 553
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETF 105
RR + FL+ + EA+ + +M +EL W + KKQE + A F
Sbjct 554 RRTRLAFLNVQAAEEALPRIVELMGRELNWD----DYKKQEQLETARKF 598
> ECU10g0870
Length=614
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 47/104 (45%), Gaps = 1/104 (0%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPI 61
E L R YG A + KK + L YL EV Y E A V D+L RL I
Sbjct 476 ERLARSYGTRALRLSSYIKKNR-KKVLSVKYSYLIEEVEYCIDNEMAVKVCDVLCNRLMI 534
Query 62 LFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETF 105
+D K + ID V + K+ GW A N+++ +A + + +
Sbjct 535 GLMDVKEAYQCIDKVLGVFKKKHGWDADRCNREEADAIRMLDKY 578
> YIL155c
Length=649
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 34/73 (46%), Gaps = 0/73 (0%)
Query 33 PYLKAEVLYACRYEQARSVSDILARRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATAN 92
P+ E+ Y+ +YE R+ D L RR FLD K + A+ +M E WS
Sbjct 575 PFTIGELKYSMQYEYCRTPLDFLLRRTRFAFLDAKEALNAVHATVKVMGDEFNWSEKKRQ 634
Query 93 KKKQEAKKYAETF 105
+ ++ + +TF
Sbjct 635 WELEKTVNFIKTF 647
> CE23209
Length=364
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 85 GWSAATANKKKQEAKKYAETFTAAPEEERPERP 117
G NKKK+E +Y T APEE+ P+RP
Sbjct 196 GRDGEDGNKKKKEEIEYTLRLTPAPEEQIPQRP 228
> At4g21630
Length=772
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 2/69 (2%)
Query 48 ARSVSDILAR--RLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETF 105
A+ D+LAR +P +F D+++G + + + + + SAAT + K AE
Sbjct 451 AKKPDDLLARYNSIPYIFTDYEIGTHILQYIRTTRSPTVRISAATTLNGQPAMTKVAEFS 510
Query 106 TAAPEEERP 114
+ P P
Sbjct 511 SRGPNSVSP 519
> 7303030
Length=700
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 44/111 (39%), Gaps = 34/111 (30%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEPLVEG-----LPYLKAEVLYACRYEQARSVSDILA 56
+HL + YG A V ++A P++ PY+ AE+ RY A+
Sbjct 490 QHLAKSYGDRAFAVSKMASLTGKRWPIIGNRIHPEFPYIDAEI----RYGAAQ------- 538
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFTA 107
EA+ + +M +ELGWS +K+ K+A F A
Sbjct 539 --------------EALPVIVDIMGEELGWS----KDEKERQIKHATEFLA 571
> At2g34360
Length=466
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 2/80 (2%)
Query 12 ARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPILFLDHKLGVE 71
A+ V + S++E ++ G + ++ Y V +A LPIL L H L +
Sbjct 321 AKLAVRVVLSFSIVESILVGTVLILIRKIWGFAYSSDPEVVSHVASMLPILALGHSL--D 378
Query 72 AIDTVCSMMAKELGWSAATA 91
+ TV S +A+ GW A
Sbjct 379 SFQTVLSGVARGCGWQKIGA 398
> Hs14589929
Length=1135
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 20/90 (22%), Positives = 42/90 (46%), Gaps = 2/90 (2%)
Query 30 EGLPYLKAEVLYACR-YEQARSVSDILARRLPILFLD-HKLGVEAIDTVCSMMAKELGWS 87
+G P L +VL C +E A SV+ + LD + + ++ +C+++ +
Sbjct 661 KGNPQLHTKVLLKCMIFEYAESVTSTAMTSVSQASLDVSMIIIISLGAICAVLLVIMVLF 720
Query 88 AATANKKKQEAKKYAETFTAAPEEERPERP 117
A N++K++ + Y + + P+RP
Sbjct 721 ATRCNREKKDTRSYNCRVAESTYQHHPKRP 750
> At2g31100
Length=355
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 1/34 (2%)
Query 5 VRHYGFMARDVVELAKKESLMEPLVEGLPYLKAE 38
+ HYG MA ++ KK SL+EP V P L E
Sbjct 33 ILHYGDMAEEIAATPKKSSLLEP-VTSKPTLSDE 65
> Hs22050467_1
Length=375
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 36/88 (40%), Gaps = 10/88 (11%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEP-----LVEGLPYLKAEV-----LYACRYEQARSV 51
+HL YG A +V +A S P LV PY++AEV Y+C S
Sbjct 262 QHLAATYGDKAFEVATMASVTSKRWPIVGVRLVSEFPYIEAEVNYGIKEYSCTTVDMISH 321
Query 52 SDILARRLPILFLDHKLGVEAIDTVCSM 79
SD +R D K + +D C +
Sbjct 322 SDRYKKRFCKSDADQKGFITIVDVPCVL 349
> 7293760
Length=576
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGW 86
R +P FLDH V I+ +C+ A E G+
Sbjct 163 REIPAEFLDHPQLVNGIEDICTTRATEDGF 192
> 7303426
Length=827
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 19/99 (19%), Positives = 47/99 (47%), Gaps = 13/99 (13%)
Query 1 REHLVRHYGFMARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLP 60
RE +++H F+ R +L +E + ++ + YL+ + + A +++ +++R+P
Sbjct 665 REEILKHCRFLVR---KLRYEEMTRQDPLKAMQYLRHHIADVIDHSDAEQLNNTISKRIP 721
Query 61 -------ILFLDHKLGVEAIDTVCSMMAKELGWSAATAN 92
+L ++++ +D + +EL AT N
Sbjct 722 MYRQRSKVLLVNNEF---VLDNSTCIFTEELADGPATEN 757
> CE18306
Length=809
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 29/73 (39%), Gaps = 2/73 (2%)
Query 45 YEQARSVSDILARRL--PILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYA 102
YE A + I R PI F + ++ V I C + + T NKKK K+
Sbjct 539 YETANDIPGIKIFRFDSPIYFGNSEMFVRKIHQACGLNPLIVRGELETENKKKDARKEKE 598
Query 103 ETFTAAPEEERPE 115
E P+E E
Sbjct 599 EEDAEIPKETEKE 611
> At4g33030
Length=477
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 26/55 (47%), Gaps = 4/55 (7%)
Query 37 AEVLYACRYEQARSVSDILARRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATA 91
A L+ + + D L RRL DH+LG+E++ + S+ + W A T
Sbjct 99 ATALHLSKKNYEVCIVDNLVRRL----FDHQLGLESLTPIASIHDRISRWKALTG 149
Lambda K H
0.318 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40