bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6172_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
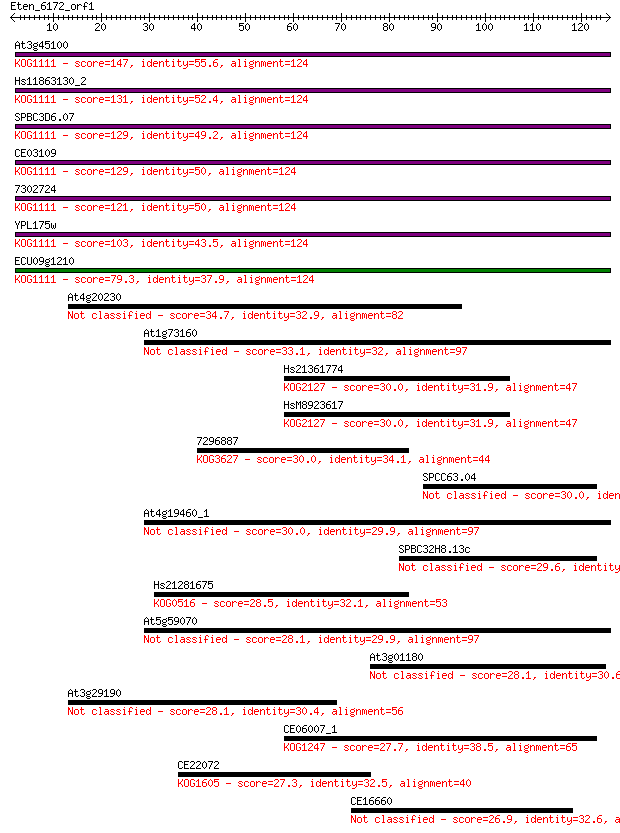
At3g45100 147 4e-36
Hs11863130_2 131 3e-31
SPBC3D6.07 129 2e-30
CE03109 129 2e-30
7302724 121 3e-28
YPL175w 103 9e-23
ECU09g1210 79.3 2e-15
At4g20230 34.7 0.041
At1g73160 33.1 0.12
Hs21361774 30.0 1.1
HsM8923617 30.0 1.2
7296887 30.0 1.2
SPCC63.04 30.0 1.2
At4g19460_1 30.0 1.2
SPBC32H8.13c 29.6 1.3
Hs21281675 28.5 3.0
At5g59070 28.1 3.7
At3g01180 28.1 4.2
At3g29190 28.1 4.5
CE06007_1 27.7 6.1
CE22072 27.3 8.0
CE16660 26.9 8.6
> At3g45100
Length=450
Score = 147 bits (371), Expect = 4e-36, Method: Composition-based stats.
Identities = 69/124 (55%), Positives = 88/124 (70%), Gaps = 1/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+GY+VV+TDHS++ FAD IHMNK+ +F L D ICVSHT KEN VLR L+P ++
Sbjct 119 MGYKVVFTDHSLYGFADVGSIHMNKVLQFSLADIDQAICVSHTSKENTVLRSGLSPAKVF 178
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
+I NAVD + P S RP I I+++SRL YRKG DLLV++IP VC YPNV F++G
Sbjct 179 MIPNAVDTAMFKP-ASVRPSTDIITIVVISRLVYRKGADLLVEVIPEVCRLYPNVRFVVG 237
Query 122 GDGP 125
GDGP
Sbjct 238 GDGP 241
> Hs11863130_2
Length=463
Score = 131 bits (330), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 65/124 (52%), Positives = 86/124 (69%), Gaps = 2/124 (1%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G + V+TDHS+F FAD + + NK+ L +H ICVS+T KEN VLR ALNP ++
Sbjct 124 MGLQTVFTDHSLFGFADVSSVLTNKLLTVSLCDTNHIICVSYTSKENTVLRAALNPEIVS 183
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NAVD + PD +R +I I+++SRL YRKGIDLL IIP +C KYP++ FIIG
Sbjct 184 VIPNAVDPTDFTPDPFRR--HDSITIVVVSRLVYRKGIDLLSGIIPELCQKYPDLNFIIG 241
Query 122 GDGP 125
G+GP
Sbjct 242 GEGP 245
> SPBC3D6.07
Length=456
Score = 129 bits (323), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 61/124 (49%), Positives = 85/124 (68%), Gaps = 1/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G + +TDHS+F FADA I NK+ KF ++ +H ICVSHT +EN VLR LNP +++
Sbjct 109 MGLKTCFTDHSLFGFADAGSIVTNKLLKFTMSDVNHVICVSHTCRENTVLRAVLNPKRVS 168
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NA+ + PD SK + I+++SRL Y KGIDLL+ +IP +CA++P V F+I
Sbjct 169 VIPNALVAENFQPDPSK-ASKDFLTIVVISRLYYNKGIDLLIAVIPRICAQHPKVRFVIA 227
Query 122 GDGP 125
GDGP
Sbjct 228 GDGP 231
> CE03109
Length=444
Score = 129 bits (323), Expect = 2e-30, Method: Composition-based stats.
Identities = 62/125 (49%), Positives = 87/125 (69%), Gaps = 2/125 (1%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNK-IFKFFLTHCDHCICVSHTHKENLVLRGALNPHQI 60
+G R V+TDHS+F FADA+ I NK + ++ L + D ICVS+T KEN VLRG L+P+++
Sbjct 120 MGLRTVFTDHSLFGFADASAILTNKLVLQYSLINVDQTICVSYTSKENTVLRGKLDPNKV 179
Query 61 AVINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFII 120
+ I NA++ + PD ++ P I+ L RL YRKG DLL +I+P VCA++ +V FII
Sbjct 180 STIPNAIETSLFTPDRNQFFNNPT-TIVFLGRLVYRKGADLLCEIVPKVCARHKSVRFII 238
Query 121 GGDGP 125
GGDGP
Sbjct 239 GGDGP 243
> 7302724
Length=479
Score = 121 bits (304), Expect = 3e-28, Method: Composition-based stats.
Identities = 62/124 (50%), Positives = 78/124 (62%), Gaps = 1/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
LG + V+TDHS+F FAD + N + + L +H ICVSH KEN VLR + H+++
Sbjct 113 LGLKTVFTDHSLFGFADLSAALTNNLLEVNLGMVNHAICVSHIGKENTVLRARVAKHRVS 172
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NAVD + PD +RP INI++ SRL YRKGIDLL IIP PN+ FII
Sbjct 173 VIPNAVDTALFTPDPQQRPSNDIINIVVASRLVYRKGIDLLAGIIPRF-KNTPNINFIIV 231
Query 122 GDGP 125
GDGP
Sbjct 232 GDGP 235
> YPL175w
Length=452
Score = 103 bits (256), Expect = 9e-23, Method: Composition-based stats.
Identities = 54/129 (41%), Positives = 78/129 (60%), Gaps = 5/129 (3%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G R V+TDHS++ F + I +NK+ F LT+ D ICVS+T KEN+++R L+P I+
Sbjct 115 MGLRTVFTDHSLYGFNNLTSIWVNKLLTFTLTNIDRVICVSNTCKENMIVRTELSPDIIS 174
Query 62 VINNAVDVPAYVP-----DLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNV 116
VI NAV + P ++ I I+++ RL KG DLL IIP VC+ + +V
Sbjct 175 VIPNAVVSEDFKPRDPTGGTKRKQSRDKIVIVVIGRLFPNKGSDLLTRIIPKVCSSHEDV 234
Query 117 TFIIGGDGP 125
FI+ GDGP
Sbjct 235 EFIVAGDGP 243
> ECU09g1210
Length=408
Score = 79.3 bits (194), Expect = 2e-15, Method: Composition-based stats.
Identities = 47/124 (37%), Positives = 70/124 (56%), Gaps = 2/124 (1%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
L + V TDHS+F I +N + + L + D CICVS+T KEN +R + +I
Sbjct 115 LNLKTVITDHSIFEVGPFENIVVNALCRLALKNIDRCICVSYTSKENTHIRTMIPLERIH 174
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NA+ + P S++ ++++SRL +RKGIDLL+ IP +C +V +I
Sbjct 175 VIPNAIVSDIFRP--SEKKHSGGKVVVVVSRLVFRKGIDLLIGAIPLICKGDRSVRIVIV 232
Query 122 GDGP 125
GDGP
Sbjct 233 GDGP 236
> At4g20230
Length=612
Score = 34.7 bits (78), Expect = 0.041, Method: Composition-based stats.
Identities = 27/93 (29%), Positives = 42/93 (45%), Gaps = 17/93 (18%)
Query 13 MFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVDV--- 69
M S +AA H+ + L D + + +H E LV G PH +A I NA+ +
Sbjct 200 MLSLYEAA--HLATTKDYIL---DEALIFTSSHLETLVATGTCPPHLLARIRNALSICQH 254
Query 70 --------PAYVPDLSKRPPPPAINILILSRLS 94
A++P S+ P N+LI +R+S
Sbjct 255 WNFEFQVSEAHIPSGSQN-PYKVCNLLIFTRIS 286
> At1g73160
Length=486
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 31/107 (28%), Positives = 48/107 (44%), Gaps = 11/107 (10%)
Query 29 KFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAYV--PDLSKR------P 80
+FF + H IC+S++ +E LV L ++ VI N VD +V P+ R
Sbjct 235 RFFPKYKQH-ICISNSAREVLVNIYQLPKRKVHVIVNGVDQTKFVYSPESGARFRAKHGI 293
Query 81 PPPAINIL--ILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDGP 125
P I+ + RL KG LL + + +P V ++ G GP
Sbjct 294 PDNGTYIVMGVSGRLVRDKGHPLLYEAFALLVKMHPKVYLLVAGSGP 340
> Hs21361774
Length=791
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 58 HQIAVINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVD 104
H I+ ++ + V L KRP PP +++ LS L RK ++ L++
Sbjct 602 HGISTVSLPNSLQEVVDPLGKRPNPPPVSVPYLSPLVLRKELESLLE 648
> HsM8923617
Length=452
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 58 HQIAVINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVD 104
H I+ ++ + V L KRP PP +++ LS L RK ++ L++
Sbjct 199 HGISTVSLPNSLQEVVDPLGKRPNPPPVSVPYLSPLVLRKELESLLE 245
> 7296887
Length=392
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 40 CVSHTHKENLVLRGALNPHQIAVINNAVDVPAYVPDLSKRPPPP 83
C+S+TH + L G + P Q I + VD P+L P P
Sbjct 17 CLSNTHTQRLPPEGRMRPLQDDSIRSPVDRDIVFPELDAGPGKP 60
> SPCC63.04
Length=1369
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Query 87 ILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGG 122
++ + R S +KGID+L D+ P + K+ N+ I+ G
Sbjct 550 LVFVGRWSVQKGIDILADLAPTLLEKF-NIQLIVVG 584
> At4g19460_1
Length=510
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 29/107 (27%), Positives = 44/107 (41%), Gaps = 11/107 (10%)
Query 29 KFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAYVPD-------LSKRPP 81
+FF + H I +S + E L + ++ VI N VD + D SK
Sbjct 247 RFFHNYAHH-IAISDSCGEMLRDVYQIPEKRVHVILNGVDENGFTSDKKLRTLFRSKLGL 305
Query 82 PPAINILILS---RLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDGP 125
P + ++L RL KG LL + + Y NV ++ G GP
Sbjct 306 PENSSAIVLGAAGRLVKDKGHPLLFEAFAKIIQTYSNVYLVVAGSGP 352
> SPBC32H8.13c
Length=2352
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 82 PPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGG 122
P A ++ + R S++KGIDL+ D+ P + ++ NV I G
Sbjct 1462 PSAQLLVFVGRWSHQKGIDLIADLAPKLLTEH-NVQLITIG 1501
> Hs21281675
Length=880
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 24/56 (42%), Gaps = 3/56 (5%)
Query 31 FLTHCDHCICVSHTHKENLVLRGALNP--HQIAVINNAVDV-PAYVPDLSKRPPPP 83
+L D C C S +HK L+ P H++ V + P S+ PPPP
Sbjct 265 YLDKHDPCRCTSLSHKPGSFLKPPAPPVQHEVRVQDGPSQTQPTMTISRSQSPPPP 320
> At5g59070
Length=519
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 29/115 (25%), Positives = 47/115 (40%), Gaps = 20/115 (17%)
Query 29 KFFLTHCDHCICVSHTHKENLVLRGALNPHQ-IAVINNAVDVPAYVPDLSKRPP------ 81
KFF + H + H +++ R + P + + +I N VD + PD+SKR
Sbjct 264 KFFQRYAHHV--ATSDHCGDVLKRIYMIPEERVHIILNGVDENVFKPDVSKRESFREKFG 321
Query 82 -------PPAINILILSRLSYRKGIDLLVDIIPAVCAK----YPNVTFIIGGDGP 125
+ + I RL KG L+ + V + NV ++ GDGP
Sbjct 322 VRSGKNKKSPLVLGIAGRLVRDKGHPLMFSALKRVFEENKEARENVVVLVAGDGP 376
> At3g01180
Length=792
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 76 LSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDG 124
L RP P I + RL ++KG+DL+ + +P + ++ +V ++ G G
Sbjct 600 LPVRPDVPLIGFI--GRLDHQKGVDLIAEAVPWMMSQ--DVQLVMLGTG 644
> At3g29190
Length=519
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 5/56 (8%)
Query 13 MFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVD 68
M SF +AA + K + D + + +H E+L GA PH I NA++
Sbjct 115 MLSFYEAANMATTKDYIL-----DEALSFTSSHLESLAANGACPPHMSRRIRNALN 165
> CE06007_1
Length=754
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 36/79 (45%), Gaps = 14/79 (17%)
Query 58 HQIAVINNAVD--VPAYVPDLSKR---PPPPAINILILSRLSYRKGIDLLVDIIPAVC-- 110
H +A I + + +P YV + P P NILI + L Y + L +II V
Sbjct 3 HDLADIKKSFEASLPGYVEKKDPKSILPQPGKRNILITAALPYVNNVPHLGNIIGCVLSA 62
Query 111 ---AKYPNV----TFIIGG 122
A+Y N+ TF +GG
Sbjct 63 DVFARYCNLRGHQTFYVGG 81
> CE22072
Length=287
Score = 27.3 bits (59), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 36 DHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAYVPD 75
+HC+CV + ++L + G +P + +++NAV AY D
Sbjct 154 EHCVCVFGNYVKDLTILGR-DPSKTMILDNAVQSFAYQLD 192
> CE16660
Length=717
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query 72 YVPDLSKRPPPPAINILILSRLS----YRKGIDLLVDIIPAVCAKYPNVT 117
+ PD+ PP P + + R+S KGID+ D++ + NVT
Sbjct 23 FEPDVLLSPPAPFFSFKYIDRISTINEIAKGIDIQADLMNGKVGEILNVT 72
Lambda K H
0.328 0.144 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1183965632
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40