bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6194_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
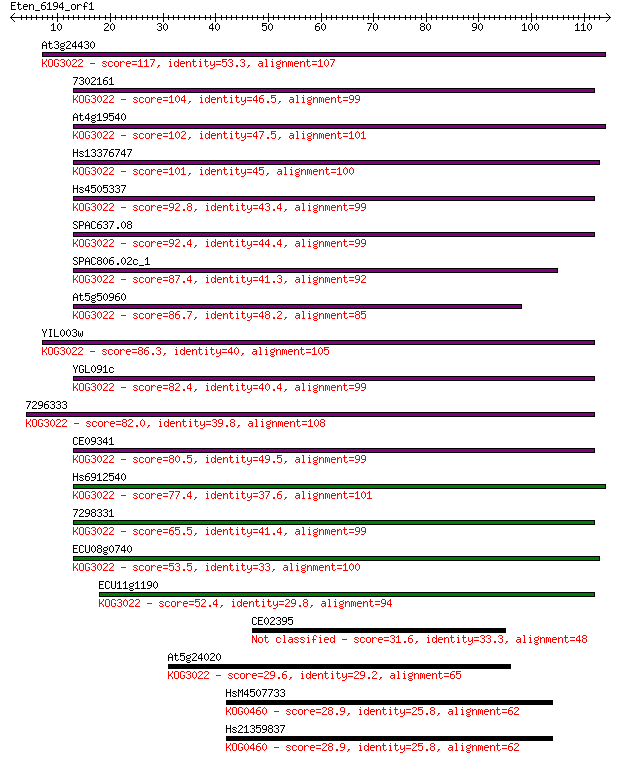
At3g24430 117 6e-27
7302161 104 3e-23
At4g19540 102 2e-22
Hs13376747 101 3e-22
Hs4505337 92.8 1e-19
SPAC637.08 92.4 2e-19
SPAC806.02c_1 87.4 6e-18
At5g50960 86.7 9e-18
YIL003w 86.3 1e-17
YGL091c 82.4 2e-16
7296333 82.0 2e-16
CE09341 80.5 7e-16
Hs6912540 77.4 7e-15
7298331 65.5 2e-11
ECU08g0740 53.5 1e-07
ECU11g1190 52.4 2e-07
CE02395 31.6 0.41
At5g24020 29.6 1.3
HsM4507733 28.9 2.2
Hs21359837 28.9 2.2
> At3g24430
Length=532
Score = 117 bits (292), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 57/107 (53%), Positives = 75/107 (70%), Gaps = 3/107 (2%)
Query 7 QGHSALRGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAPLDGCVVVTT 66
QG + +RGP VS V+ Q+LT T WG LDYLV+D+PPGTGDI +TL Q APL V+VTT
Sbjct 258 QGRAIMRGPMVSGVINQLLTTTEWGELDYLVIDMPPGTGDIQLTLC-QVAPLTAAVIVTT 316
Query 67 PQALSLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELFGR 113
PQ L+ +D KG+RM + +P +AVV NM +F D+ KR+ FG+
Sbjct 317 PQKLAFIDVAKGVRMFSKLKVPCVAVVENMCHF--DADGKRYYPFGK 361
> 7302161
Length=293
Score = 104 bits (260), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 46/99 (46%), Positives = 67/99 (67%), Gaps = 1/99 (1%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAPLDGCVVVTTPQALSL 72
RGP V S +Q++L GT WG+LD LV+D PPGTGD+H++L+ Q AP+ G ++VTTP ++
Sbjct 127 RGPLVMSAIQRLLKGTDWGLLDVLVIDTPPGTGDVHLSLS-QHAPITGVILVTTPHTAAV 185
Query 73 VDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
KG M + + +P VV NM Y +C +C++R E F
Sbjct 186 QVTLKGASMYEKLNVPIFGVVENMKYTICQNCNQRLEFF 224
> At4g19540
Length=313
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 48/101 (47%), Positives = 66/101 (65%), Gaps = 1/101 (0%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAPLDGCVVVTTPQALSL 72
RGP V S + +M G WG LD LVVD+PPGTGD I+++Q L G V+V+TPQ ++L
Sbjct 132 RGPMVMSALAKMTKGVDWGDLDILVVDMPPGTGDAQISISQN-LKLSGAVIVSTPQDVAL 190
Query 73 VDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELFGR 113
DA +GI M V +P L +V NM+ FVC C++ +FG+
Sbjct 191 ADANRGISMFDKVRVPILGLVENMSCFVCPHCNEPSFIFGK 231
> Hs13376747
Length=289
Score = 101 bits (252), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 45/100 (45%), Positives = 67/100 (67%), Gaps = 1/100 (1%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAPLDGCVVVTTPQALSL 72
RG V S ++++L WG LDYLVVD+PPGTGD+ ++++Q P+ G V+V+TPQ ++L
Sbjct 127 RGLMVMSAIEKLLRQVDWGQLDYLVVDMPPGTGDVQLSVSQN-IPITGAVIVSTPQDIAL 185
Query 73 VDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELFG 112
+DA KG M + V +P L +V NM+ F C C + +FG
Sbjct 186 MDAHKGAEMFRRVHVPVLGLVQNMSVFQCPKCKHKTHIFG 225
> Hs4505337
Length=320
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/101 (42%), Positives = 63/101 (62%), Gaps = 2/101 (1%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQ--QQAPLDGCVVVTTPQAL 70
RGP + +++Q L WG +DYL+VD PPGT D H+++ + A +DG V++TTPQ L
Sbjct 145 RGPKKNGMIKQFLRDVDWGEVDYLIVDTPPGTSDEHLSVVRYLATAHIDGAVIITTPQEL 204
Query 71 SLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
SL D K I + V +P + VV NM+ F+C C K ++F
Sbjct 205 SLQDVRKEINFCRKVKLPIIGVVENMSPFICPKCKKESQIF 245
> SPAC637.08
Length=317
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/101 (43%), Positives = 63/101 (62%), Gaps = 2/101 (1%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQ--QQAPLDGCVVVTTPQAL 70
RGP + +++Q + +W LDYL+VD PPGT D H++L Q + + +DG VVVTTPQ +
Sbjct 151 RGPKKNGLIKQFIKDVNWENLDYLIVDTPPGTSDEHLSLVQFFKNSGIDGAVVVTTPQEV 210
Query 71 SLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
+L D K I + IP L +V NM+ FVC SC + +F
Sbjct 211 ALQDVRKEIDFCRKASIPILGLVENMSGFVCPSCKGKSNIF 251
> SPAC806.02c_1
Length=286
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 38/101 (37%), Positives = 62/101 (61%), Gaps = 9/101 (8%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQ---------QQAPLDGCVV 63
RGP +++++Q ++ WG LD+L++D PPGTGD H+T+ + + P+DG V+
Sbjct 104 RGPKKAAMIRQFISDVSWGELDFLIIDTPPGTGDEHLTIVESLLSETSTVRDVPIDGAVI 163
Query 64 VTTPQALSLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSC 104
VTTPQ ++ +D +K I K I L +V NM+ ++C C
Sbjct 164 VTTPQGIATLDVQKEIDFCKKASIKILGIVENMSGYICPHC 204
> At5g50960
Length=295
Score = 86.7 bits (213), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 41/87 (47%), Positives = 57/87 (65%), Gaps = 2/87 (2%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAP--LDGCVVVTTPQAL 70
RGP + +++Q L +WG +DYLVVD PPGT D HI++ Q P +DG ++VTTPQ +
Sbjct 149 RGPRKNGLIKQFLKDVYWGEIDYLVVDAPPGTSDEHISIVQYLLPTGIDGAIIVTTPQEV 208
Query 71 SLVDAEKGIRMLKSVCIPTLAVVCNMA 97
SL+D K + K V +P L VV NM+
Sbjct 209 SLIDVRKEVSFCKKVGVPVLGVVENMS 235
> YIL003w
Length=293
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 42/108 (38%), Positives = 69/108 (63%), Gaps = 3/108 (2%)
Query 7 QGHSAL-RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQ--QAPLDGCVV 63
+G+S + RGP +S+++Q ++ WG LDYL++D PPGT D HI++ ++ + DG +V
Sbjct 104 RGNSVIWRGPKKTSMIKQFISDVAWGELDYLLIDTPPGTSDEHISIAEELRYSKPDGGIV 163
Query 64 VTTPQALSLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
VTTPQ+++ D +K I K V + L ++ NM+ FVC C + +F
Sbjct 164 VTTPQSVATADVKKEINFCKKVDLKILGIIENMSGFVCPHCAECTNIF 211
> YGL091c
Length=328
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 62/101 (61%), Gaps = 2/101 (1%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQ--QQAPLDGCVVVTTPQAL 70
RG + ++++ L W LDYLV+D PPGT D HI++ + +++ +DG +VVTTPQ +
Sbjct 163 RGSKKNLLIKKFLKDVDWDKLDYLVIDTPPGTSDEHISINKYMRESGIDGALVVTTPQEV 222
Query 71 SLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
+L+D K I K I L +V NM+ FVC +C ++F
Sbjct 223 ALLDVRKEIDFCKKAGINILGLVENMSGFVCPNCKGESQIF 263
> 7296333
Length=260
Score = 82.0 bits (201), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 61/110 (55%), Gaps = 2/110 (1%)
Query 4 KNYQGHSALRGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQ--QQAPLDGC 61
KN + RGP + +++Q LT W LDYL++D PPGT D HIT+ + ++ G
Sbjct 89 KNREDPVIWRGPKKTMMIRQFLTDVRWDELDYLIIDTPPGTSDEHITVMECLKEVGCHGA 148
Query 62 VVVTTPQALSLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
++VTTPQ ++L D K I K I L +V NM+ FVC C +F
Sbjct 149 IIVTTPQEVALDDVRKEITFCKKTGINILGIVENMSGFVCPHCTSCTNIF 198
> CE09341
Length=334
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 49/123 (39%), Positives = 65/123 (52%), Gaps = 24/123 (19%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQ---QQAPLDGCVVVTTPQA 69
RG + +++Q L WG +DYL++D PPGT D HI+L Q Q PLDG ++V+TPQ
Sbjct 149 RGARKNGMIKQFLKDVDWGEVDYLLIDTPPGTSDEHISLVQFLLQAGPLDGALIVSTPQE 208
Query 70 LSLVDAEKGIRM-LK----------------SVC----IPTLAVVCNMAYFVCDSCDKRH 108
+SL+D K + LK S C +P L VV NMA FVC +C
Sbjct 209 VSLLDVRKEASLNLKLSLKKSFQNKFLILTVSFCVKTKVPILGVVENMARFVCPNCAHTT 268
Query 109 ELF 111
LF
Sbjct 269 LLF 271
> Hs6912540
Length=271
Score = 77.4 bits (189), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 59/103 (57%), Gaps = 2/103 (1%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAPLD--GCVVVTTPQAL 70
RGP +++++Q ++ WG LDYLVVD PPGT D H+ + P G +VVTTPQA+
Sbjct 106 RGPKKNALIKQFVSDVAWGELDYLVVDTPPGTSDEHMATIEALRPYQPLGALVVTTPQAV 165
Query 71 SLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELFGR 113
S+ D + + + + + +V NM+ F C C + +F R
Sbjct 166 SVGDVRRELTFCRKTGLRVMGIVENMSGFTCPHCTECTSVFSR 208
> 7298331
Length=311
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 57/104 (54%), Gaps = 5/104 (4%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHIT----LTQQQAPLD-GCVVVTTP 67
RGP + +++Q L+ WG LD L++D PPGT D H++ L P V+VTTP
Sbjct 147 RGPKKNGMIRQFLSEVDWGNLDLLLLDTPPGTSDEHLSVVSYLKDDANPESLRAVMVTTP 206
Query 68 QALSLVDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
Q +SL+D K I K IP + V+ NM+ F C C E+F
Sbjct 207 QEVSLLDVRKEINFCKKQNIPIVGVIENMSSFRCGHCGNSSEIF 250
> ECU08g0740
Length=239
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 48/100 (48%), Gaps = 5/100 (5%)
Query 13 RGPFVSSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAPLDGCVVVTTPQALSL 72
RGP SV+ D +V D+PPG + H L + G +++TTPQ +SL
Sbjct 88 RGPKKMSVLSMFYESIDG--FDNVVFDMPPGISEEHGFLIGKDV---GALIITTPQNVSL 142
Query 73 VDAEKGIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELFG 112
D+ K I S I L +V NM+ + C+ C +FG
Sbjct 143 GDSSKAIDFCASNGIRILGLVENMSGYCCECCGSSVNIFG 182
> ECU11g1190
Length=292
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 45/94 (47%), Gaps = 1/94 (1%)
Query 18 SSVVQQMLTGTHWGILDYLVVDLPPGTGDIHITLTQQQAPLDGCVVVTTPQALSLVDAEK 77
+S ++++L + D L++D PP D H+ + P G +VVTTPQ SL D +
Sbjct 144 TSAMKKLLKWCSYEGTDVLLLDTPPNVTDEHLGMVNFIRPRFG-IVVTTPQKFSLQDVAR 202
Query 78 GIRMLKSVCIPTLAVVCNMAYFVCDSCDKRHELF 111
+ + I L ++ NM F C C +F
Sbjct 203 QVDFCRKARIEVLGIIENMKRFTCQKCGHSKSIF 236
> CE02395
Length=400
Score = 31.6 bits (70), Expect = 0.41, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 47 IHITLTQQQAPLDGCVVVTTPQALSLVDAEKGIRMLKSVCIPTLAVVC 94
I + T QA LDG +++PQ SLVD +++K + L++ C
Sbjct 190 ITVACTTNQAALDGLAQISSPQ-FSLVDEMNTSKLVKQLTTALLSINC 236
> At5g24020
Length=326
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Query 31 GILDYLVVDLPPGTGDIHITLTQQQAPLDGCVVVTTPQALSLVDAEKGIRMLKSVCIPTL 90
G D++++D P G IT P + V+VTTP +L DA++ +L+ I +
Sbjct 173 GSPDFIIIDCPAGIDAGFITAI---TPANEAVLVTTPDITALRDADRVTGLLECDGIRDI 229
Query 91 AVVCN 95
++ N
Sbjct 230 KMIVN 234
> HsM4507733
Length=452
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 27/62 (43%), Gaps = 0/62 (0%)
Query 42 PGTGDIHITLTQQQAPLDGCVVVTTPQALSLVDAEKGIRMLKSVCIPTLAVVCNMAYFVC 101
PG D + APLDGC++V + + + + + + + + V N A V
Sbjct 128 PGHADYVKNMITGTAPLDGCILVVAANDGPMPQTREHLLLARQIGVEHVVVYVNKADAVQ 187
Query 102 DS 103
DS
Sbjct 188 DS 189
> Hs21359837
Length=452
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 27/62 (43%), Gaps = 0/62 (0%)
Query 42 PGTGDIHITLTQQQAPLDGCVVVTTPQALSLVDAEKGIRMLKSVCIPTLAVVCNMAYFVC 101
PG D + APLDGC++V + + + + + + + + V N A V
Sbjct 128 PGHADYVKNMITGTAPLDGCILVVAANDGPMPQTREHLLLARQIGVEHVVVYVNKADAVQ 187
Query 102 DS 103
DS
Sbjct 188 DS 189
Lambda K H
0.324 0.139 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181971906
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40