bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6205_orf3
Length=69
Score E
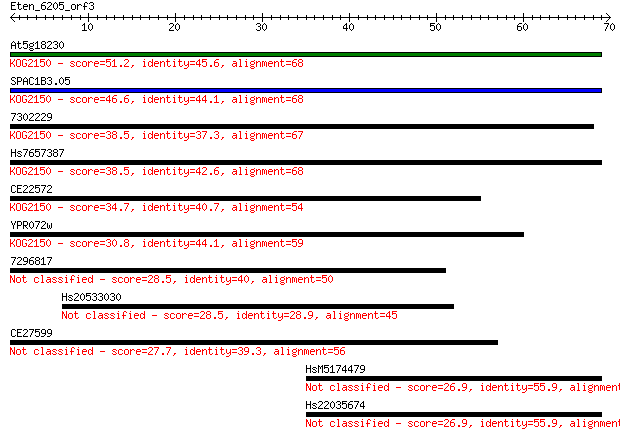
Sequences producing significant alignments: (Bits) Value
At5g18230 51.2 5e-07
SPAC1B3.05 46.6 1e-05
7302229 38.5 0.003
Hs7657387 38.5 0.003
CE22572 34.7 0.047
YPR072w 30.8 0.62
7296817 28.5 3.6
Hs20533030 28.5 3.6
CE27599 27.7 5.8
HsM5174479 26.9 8.6
Hs22035674 26.9 8.9
> At5g18230
Length=889
Score = 51.2 bits (121), Expect = 5e-07, Method: Composition-based stats.
Identities = 31/73 (42%), Positives = 44/73 (60%), Gaps = 5/73 (6%)
Query 1 LEDSRKKIERCMETFRQCEKEIKTKAFSKEGLAAKTR-----DGEDRHREALQLLQQQLQ 55
L D+RK IE+ ME F+ CEKE KTKAFSKEGL + + + R+ L + +L+
Sbjct 84 LVDARKLIEKEMERFKICEKETKTKAFSKEGLGQQPKTDPKEKAKSETRDWLNNVVSELE 143
Query 56 REAALLEAEMDGL 68
+ EAE++GL
Sbjct 144 SQIDSFEAELEGL 156
> SPAC1B3.05
Length=630
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 47/73 (64%), Gaps = 5/73 (6%)
Query 1 LEDSRKKIERCMETFRQCEKEIKTKAFSKEGL--AAKTRDGEDRHREALQLLQ---QQLQ 55
L ++R+ IE ME F+ E+E+K KAFSKEGL A+K E ++ +Q + ++L+
Sbjct 66 LLENRRLIEAKMEEFKAVEREMKIKAFSKEGLSIASKLDPKEKEKQDTIQWISNAVEELE 125
Query 56 REAALLEAEMDGL 68
R+A L+EAE + L
Sbjct 126 RQAELIEAEAESL 138
> 7302229
Length=948
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 9/76 (11%)
Query 1 LEDSRKKIERCMETFRQCEKEIKTKAFSKEGLAAKTR---------DGEDRHREALQLLQ 51
L ++R+ IE ME F+ E+E KTKA+SKEGL A + D + ++ LQ
Sbjct 82 LLENRRLIETQMERFKVVERETKTKAYSKEGLGAAQKMDPAQRIKDDARNWLTSSISSLQ 141
Query 52 QQLQREAALLEAEMDG 67
Q+ + + +E+ + G
Sbjct 142 IQIDQYESEIESLLAG 157
> Hs7657387
Length=753
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 29/73 (39%), Positives = 38/73 (52%), Gaps = 5/73 (6%)
Query 1 LEDSRKKIERCMETFRQCEKEIKTKAFSKE--GLAAKTRDGEDRHREALQLLQQ---QLQ 55
L D+RK IE ME F+ E+E KTKA+SKE GLA K + E Q L L
Sbjct 77 LIDNRKLIETQMERFKVVERETKTKAYSKEGLGLAQKVDPAQKEKEEVGQWLTNTIDTLN 136
Query 56 REAALLEAEMDGL 68
+ E+E++ L
Sbjct 137 MQVDQFESEVESL 149
> CE22572
Length=513
Score = 34.7 bits (78), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 35/56 (62%), Gaps = 2/56 (3%)
Query 1 LEDSRKKIERCMETFRQCEKEIKTKAFSKEGLAAKTR-DGEDRHR-EALQLLQQQL 54
L RK IE+ ME F+ E+E KTK SK GL+A+ + D +++ + E + +Q Q+
Sbjct 77 LNSYRKLIEQRMEQFKDVERENKTKPHSKLGLSAEEKLDPKEKEKAETMDWIQHQI 132
> YPR072w
Length=560
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 26/69 (37%), Positives = 34/69 (49%), Gaps = 10/69 (14%)
Query 1 LEDSRKKIERCMETFRQCEKEIKTKAFSKEGLA----------AKTRDGEDRHREALQLL 50
L +R+ IE ME F+ EK +KTK FSKE L K RD + L L
Sbjct 79 LMTNRRLIENGMERFKSVEKLMKTKQFSKEALTNPDIIKDPKELKKRDQVLFIHDCLDEL 138
Query 51 QQQLQREAA 59
Q+QL++ A
Sbjct 139 QKQLEQYEA 147
> 7296817
Length=967
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 5/55 (9%)
Query 1 LEDSRKKIERCMETFRQCEKEIKTKAFSKEGLAAKTRD-----GEDRHREALQLL 50
L+D R K++R R +EIK + FS+E LA RD E R+ +AL ++
Sbjct 808 LDDQRMKLDRANREIRTQLREIKARPFSEEYLAQFERDLSLQELEARNTKALNMI 862
> Hs20533030
Length=544
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 22/45 (48%), Gaps = 0/45 (0%)
Query 7 KIERCMETFRQCEKEIKTKAFSKEGLAAKTRDGEDRHREALQLLQ 51
KI RC+ + +++ TK + E A +D E HR L++
Sbjct 282 KISRCLPSPDSGLEDLSTKCLTVEACRASAQDAEQPHRPLPSLME 326
> CE27599
Length=305
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 39/68 (57%), Gaps = 12/68 (17%)
Query 1 LEDSRKK--IERCMETFRQCEKEI----KTKAFS------KEGLAAKTRDGEDRHREALQ 48
L DSR+ IE C++ +Q + +I K KA S +G + +TR G+D+ RE ++
Sbjct 162 LRDSRQASIIESCVDFQKQIQWKIGIMEKVKAKSGRSYPMSDGPSVRTRGGQDKQREEMK 221
Query 49 LLQQQLQR 56
Q++L++
Sbjct 222 KQQKELEK 229
> HsM5174479
Length=3321
Score = 26.9 bits (58), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 24/36 (66%), Gaps = 2/36 (5%)
Query 35 KTRDGEDRHREALQLLQQQLQ--REAALLEAEMDGL 68
K RD E ++E L LLQQ+LQ RE ALL++ GL
Sbjct 526 KLRDAEKTYQEDLTLLQQRLQGAREDALLDSVEVGL 561
> Hs22035674
Length=3336
Score = 26.9 bits (58), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 24/36 (66%), Gaps = 2/36 (5%)
Query 35 KTRDGEDRHREALQLLQQQLQ--REAALLEAEMDGL 68
K RD E ++E L LLQQ+LQ RE ALL++ GL
Sbjct 526 KLRDAEKTYQEDLTLLQQRLQGAREDALLDSVEVGL 561
Lambda K H
0.314 0.129 0.344
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200381448
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40