bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6333_orf2
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
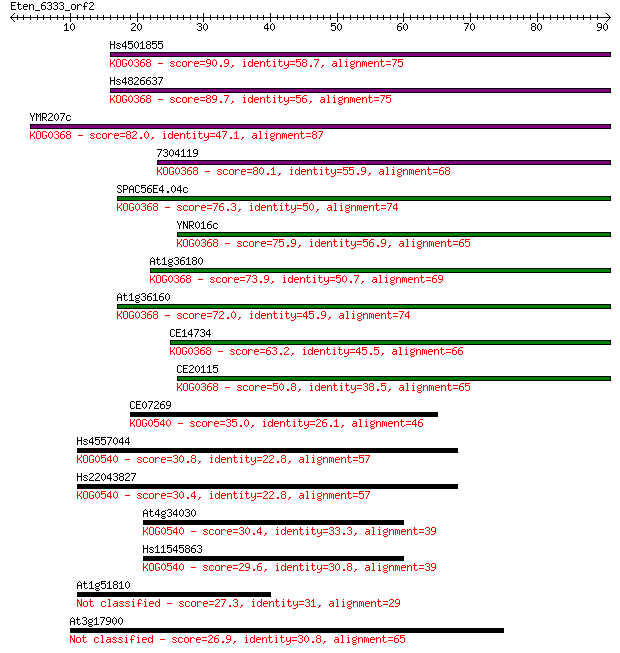
Hs4501855 90.9 5e-19
Hs4826637 89.7 1e-18
YMR207c 82.0 3e-16
7304119 80.1 9e-16
SPAC56E4.04c 76.3 1e-14
YNR016c 75.9 2e-14
At1g36180 73.9 7e-14
At1g36160 72.0 2e-13
CE14734 63.2 1e-10
CE20115 50.8 5e-07
CE07269 35.0 0.035
Hs4557044 30.8 0.72
Hs22043827 30.4 0.75
At4g34030 30.4 0.94
Hs11545863 29.6 1.4
At1g51810 27.3 7.6
At3g17900 26.9 9.6
> Hs4501855
Length=2483
Score = 90.9 bits (224), Expect = 5e-19, Method: Composition-based stats.
Identities = 44/75 (58%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + W SGF D GSFKE+ PW VV GRARLGG P+GVIAV+ RT PADPA
Sbjct 2079 HPTLKGTWQSGFFDHGSFKEIMAPWAQTVVTGRARLGGIPVGVIAVETRTVEVAVPADPA 2138
Query 76 NADSARVEIPQAGQV 90
N DS I QAGQV
Sbjct 2139 NLDSEAKIIQQAGQV 2153
> Hs4826637
Length=2346
Score = 89.7 bits (221), Expect = 1e-18, Method: Composition-based stats.
Identities = 42/75 (56%), Positives = 49/75 (65%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + +W+SGF D GSF E+ PW VVVGRARLGG P+GV+AV+ RT PADPA
Sbjct 1943 HPTQKGQWLSGFFDYGSFSEIMQPWAQTVVVGRARLGGIPVGVVAVETRTVELSVPADPA 2002
Query 76 NADSARVEIPQAGQV 90
N DS I AGQV
Sbjct 2003 NLDSEAKIIQHAGQV 2017
> YMR207c
Length=2123
Score = 82.0 bits (201), Expect = 3e-16, Method: Composition-based stats.
Identities = 41/93 (44%), Positives = 55/93 (59%), Gaps = 6/93 (6%)
Query 4 DAEGAAEVPQQQHKKHQRRW------VSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIG 57
D + A +VP + + +W SG D+ SF E W GV+VGRARLGG P+G
Sbjct 1727 DFKPAKQVPYEARWLIEGKWDSNNNFQSGLFDKDSFFETLSGWAKGVIVGRARLGGIPVG 1786
Query 58 VIAVDPRTTTAVAPADPANADSARVEIPQAGQV 90
VIAV+ +T + PADPAN DS+ + +AGQV
Sbjct 1787 VIAVETKTIEEIIPADPANLDSSEFSVKEAGQV 1819
> 7304119
Length=2348
Score = 80.1 bits (196), Expect = 9e-16, Method: Composition-based stats.
Identities = 38/68 (55%), Positives = 44/68 (64%), Gaps = 0/68 (0%)
Query 23 WVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARV 82
W +GF DR S+ E+ W VV GRARLGG P+GVIAV+ RT PADPAN DS
Sbjct 1947 WENGFFDRDSWSEIMASWAKTVVTGRARLGGVPVGVIAVETRTVEVEMPADPANLDSEAK 2006
Query 83 EIPQAGQV 90
+ QAGQV
Sbjct 2007 TLQQAGQV 2014
> SPAC56E4.04c
Length=2280
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 37/74 (50%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 17 KKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPAN 76
K+ + ++ G D+GSF+E W VVVGRAR+GG P GVIAV+ RT PADPAN
Sbjct 1890 KEDEDSFLYGLFDKGSFQETLNGWAKTVVVGRARMGGIPTGVIAVETRTIENTVPADPAN 1949
Query 77 ADSARVEIPQAGQV 90
DS + +AGQV
Sbjct 1950 PDSTEQVLMEAGQV 1963
> YNR016c
Length=2233
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/65 (56%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 26 GFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARVEIP 85
G D+GSF E W GVVVGRARLGG P+GVI V+ RT + PADPAN +SA I
Sbjct 1859 GLFDKGSFFETLSGWAKGVVVGRARLGGIPLGVIGVETRTVENLIPADPANPNSAETLIQ 1918
Query 86 QAGQV 90
+ GQV
Sbjct 1919 EPGQV 1923
> At1g36180
Length=2359
Score = 73.9 bits (180), Expect = 7e-14, Method: Composition-based stats.
Identities = 35/69 (50%), Positives = 43/69 (62%), Gaps = 0/69 (0%)
Query 22 RWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSAR 81
+W+ G D+ SF E W VV GRA+LGG PIGV+AV+ +T V PADP DS
Sbjct 1968 KWLGGIFDKNSFVETLEGWARTVVTGRAKLGGIPIGVVAVETQTVMHVIPADPGQLDSHE 2027
Query 82 VEIPQAGQV 90
+PQAGQV
Sbjct 2028 RVVPQAGQV 2036
> At1g36160
Length=2247
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 17 KKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPAN 76
K + +W+ G D+ SF E W VV GRA+LGG P+GV+AV+ +T + PADP
Sbjct 1851 KDNTGKWLGGIFDKNSFIETLEGWARTVVTGRAKLGGIPVGVVAVETQTVMQIIPADPGQ 1910
Query 77 ADSARVEIPQAGQV 90
DS +PQAGQV
Sbjct 1911 LDSHERVVPQAGQV 1924
> CE14734
Length=2054
Score = 63.2 bits (152), Expect = 1e-10, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 39/66 (59%), Gaps = 0/66 (0%)
Query 25 SGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARVEI 84
+G CD SF E+ G W +V GRARL G PIGV++ + R + + PADPA S
Sbjct 1705 TGICDTMSFDEICGDWAKSIVAGRARLCGIPIGVVSSEFRNFSTIVPADPAIDGSQVQNT 1764
Query 85 PQAGQV 90
+AGQV
Sbjct 1765 QRAGQV 1770
> CE20115
Length=1657
Score = 50.8 bits (120), Expect = 5e-07, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 0/65 (0%)
Query 26 GFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARVEIP 85
G D SF E++ W A +V GRA+L G P+GVIA ++ + AD + +S +
Sbjct 1385 GLFDTDSFDEIRSSWAASMVTGRAKLNGLPVGVIASQWKSYEKLLLADESVENSEEMITS 1444
Query 86 QAGQV 90
+AGQV
Sbjct 1445 KAGQV 1449
> CE07269
Length=536
Score = 35.0 bits (79), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 12/46 (26%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 19 HQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPR 64
+ + V D G F E+ + +V+G AR+ G+ +G++ +P+
Sbjct 311 NMKDVVHALVDEGDFFEIMPDYAKNLVIGFARMNGRTVGIVGNNPK 356
> Hs4557044
Length=539
Score = 30.8 bits (68), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 13/58 (22%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 11 VPQQQHKKHQR-RWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTT 67
VP + K + + D F E+ + ++VG AR+ G+ +G++ P+ +
Sbjct 306 VPLESTKAYNMVDIIHSVVDEREFFEIMPNYAKNIIVGFARMNGRTVGIVGNQPKVAS 363
> Hs22043827
Length=539
Score = 30.4 bits (67), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 13/58 (22%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 11 VPQQQHKKHQR-RWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTT 67
VP + K + + D F E+ + ++VG AR+ G+ +G++ P+ +
Sbjct 306 VPLESTKAYNMVDIIHSVVDEREFFEIMPNYAKNIIVGFARMNGRTVGIVGNQPKVAS 363
> At4g34030
Length=630
Score = 30.4 bits (67), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 21 RRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVI 59
R ++ D F E + +G +V G AR+ GQ +G+I
Sbjct 370 RSIIARIVDGSEFDEFKKQYGTTLVTGFARIYGQTVGII 408
> Hs11545863
Length=563
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 21 RRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVI 59
R ++ D F E + +G +V G AR+ G P+G++
Sbjct 332 REVIARIVDGSRFTEFKAFYGDTLVTGFARIFGYPVGIV 370
> At1g51810
Length=843
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 9/29 (31%), Positives = 14/29 (48%), Gaps = 0/29 (0%)
Query 11 VPQQQHKKHQRRWVSGFCDRGSFKEVQGP 39
+ Q + K+H WV G +G K + P
Sbjct 751 IDQNREKRHIAEWVGGMLTKGDIKSITDP 779
> At3g17900
Length=838
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 29/66 (43%), Gaps = 5/66 (7%)
Query 10 EVPQQQHKKHQRRWVSGFCDRGSFK-EVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTA 68
E+ +++ + R VS F DRG F V G GV +A+ P G I V +
Sbjct 259 EMDEERKRTKSRPCVSAFVDRGDFDPNVSG----GVARSKAKCCALPNGDIVVSLQVYIV 314
Query 69 VAPADP 74
P +P
Sbjct 315 DCPKEP 320
Lambda K H
0.315 0.132 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1177758614
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40