bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6364_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
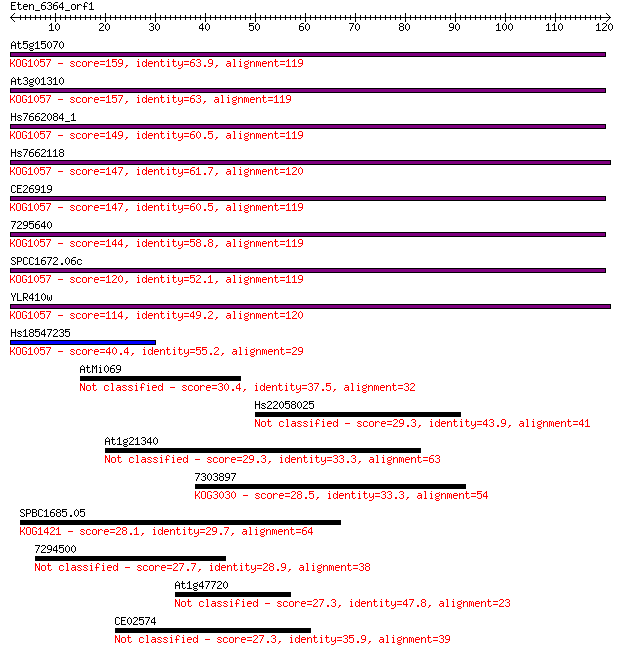
At5g15070 159 1e-39
At3g01310 157 5e-39
Hs7662084_1 149 9e-37
Hs7662118 147 3e-36
CE26919 147 4e-36
7295640 144 3e-35
SPCC1672.06c 120 4e-28
YLR410w 114 4e-26
Hs18547235 40.4 8e-04
AtMi069 30.4 0.77
Hs22058025 29.3 1.7
At1g21340 29.3 2.1
7303897 28.5 3.3
SPBC1685.05 28.1 3.6
7294500 27.7 4.9
At1g47720 27.3 7.0
CE02574 27.3 7.3
> At5g15070
Length=1030
Score = 159 bits (402), Expect = 1e-39, Method: Composition-based stats.
Identities = 76/119 (63%), Positives = 92/119 (77%), Gaps = 0/119 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
EKP D+H IYYP +AGGG K+LFRK N SS + PD+ +VR++G Y+YEEF PT G
Sbjct 142 EKPVNGDDHSIMIYYPSSAGGGMKELFRKVGNRSSEFHPDVRRVRREGSYIYEEFMPTGG 201
Query 61 TDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQ 119
TDVKVYTVGP +AHAEARKSP VDGVV R+ DGKEVRYPV+LT EK +A ++ AF+Q
Sbjct 202 TDVKVYTVGPEYAHAEARKSPVVDGVVMRNPDGKEVRYPVLLTPAEKQMAREVCIAFRQ 260
> At3g01310
Length=1015
Score = 157 bits (397), Expect = 5e-39, Method: Composition-based stats.
Identities = 75/119 (63%), Positives = 91/119 (76%), Gaps = 0/119 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
EKP D+H IYYP +AGGG K+LFRK N SS + PD+ +VR++G Y+YEEF T G
Sbjct 125 EKPVNGDDHSIMIYYPSSAGGGMKELFRKIGNRSSEFHPDVRRVRREGSYIYEEFMATGG 184
Query 61 TDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQ 119
TDVKVYTVGP +AHAEARKSP VDGVV R+ DGKEVRYPV+LT EK +A ++ AF+Q
Sbjct 185 TDVKVYTVGPEYAHAEARKSPVVDGVVMRNTDGKEVRYPVLLTPAEKQMAREVCIAFRQ 243
> Hs7662084_1
Length=1099
Score = 149 bits (377), Expect = 9e-37, Method: Composition-based stats.
Identities = 72/119 (60%), Positives = 93/119 (78%), Gaps = 1/119 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
EKP A++H+ +IYYP +AGGG ++LFRK + SS Y P+ VRK G Y+YEEF PT G
Sbjct 197 EKPVSAEDHNVYIYYPSSAGGGSQRLFRKIGSRSSVYSPE-SSVRKTGSYIYEEFMPTDG 255
Query 61 TDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQ 119
TDVKVYTVGP +AHAEARKSP++DG V R ++GKE+RYPV+LT +EK +A K+ AF+Q
Sbjct 256 TDVKVYTVGPDYAHAEARKSPALDGKVERDSEGKEIRYPVMLTAMEKLVARKVCVAFKQ 314
> Hs7662118
Length=1243
Score = 147 bits (372), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 74/120 (61%), Positives = 92/120 (76%), Gaps = 1/120 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
EKP A++H+ +IYYP +AGGG ++LFRK + SS Y P+ VRK G Y+YEEF PT G
Sbjct 186 EKPVSAEDHNVYIYYPTSAGGGSQRLFRKIGSRSSVYSPE-SNVRKTGSYIYEEFMPTDG 244
Query 61 TDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQV 120
TDVKVYTVGP +AHAEARKSP++DG V R ++GKEVRYPVIL EK IA+K+ AF+Q
Sbjct 245 TDVKVYTVGPDYAHAEARKSPALDGKVERDSEGKEVRYPVILNAREKLIAWKVCLAFKQT 304
> CE26919
Length=1339
Score = 147 bits (372), Expect = 4e-36, Method: Composition-based stats.
Identities = 72/119 (60%), Positives = 89/119 (74%), Gaps = 1/119 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
EKP +++H+ +IYYP + GGG ++LFRK N SS Y P +VRK+G Y+YEEF P G
Sbjct 161 EKPISSEDHNVYIYYPSSVGGGSQRLFRKINNRSSWYSPK-SEVRKEGSYIYEEFIPADG 219
Query 61 TDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQ 119
TDVKVY VGP +AHAEARK+P +DG V R +DGKEVRYPVIL+ EK IA K+V AF Q
Sbjct 220 TDVKVYAVGPFYAHAEARKAPGLDGKVERDSDGKEVRYPVILSNKEKQIAKKIVLAFGQ 278
> 7295640
Length=1751
Score = 144 bits (364), Expect = 3e-35, Method: Composition-based stats.
Identities = 70/119 (58%), Positives = 92/119 (77%), Gaps = 1/119 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
EKP A++H+ +IYYP +AGGG ++LFRK + SS Y P+ +VRK G ++YE+F PT G
Sbjct 209 EKPVSAEDHNIYIYYPTSAGGGSQRLFRKIGSRSSVYSPE-SRVRKTGSFIYEDFMPTDG 267
Query 61 TDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQ 119
TDVKVYTVGP +AHAEARKSP++DG V R ++GKE+RYPVIL EK I+ K+ AF+Q
Sbjct 268 TDVKVYTVGPDYAHAEARKSPALDGKVERDSEGKEIRYPVILNHSEKLISRKVCLAFKQ 326
> SPCC1672.06c
Length=920
Score = 120 bits (302), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 62/120 (51%), Positives = 83/120 (69%), Gaps = 1/120 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPT-F 59
EKP ++H+ +IY+PK+ GGG +KLFRK N SS Y PD+ R +G ++YEEF
Sbjct 196 EKPVYGEDHNIYIYFPKSVGGGGRKLFRKVANKSSDYDPDLCAPRTEGSFIYEEFMNVDN 255
Query 60 GTDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQ 119
DVKVYTVGP ++HAE RKSP VDG+V R+ GKE+R+ L+E EK +A K+ AF+Q
Sbjct 256 AEDVKVYTVGPHYSHAETRKSPVVDGIVRRNPHGKEIRFITNLSEEEKNMASKISIAFEQ 315
> YLR410w
Length=1146
Score = 114 bits (286), Expect = 4e-26, Method: Composition-based stats.
Identities = 59/121 (48%), Positives = 79/121 (65%), Gaps = 1/121 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
EKP + ++H+ +IYY GGG ++LFRK N SS + P + R +G Y+YE+F T
Sbjct 350 EKPVDGEDHNIYIYYHSKNGGGGRRLFRKVGNKSSEFDPTLVHPRTEGSYIYEQFMDTDN 409
Query 61 -TDVKVYTVGPLFAHAEARKSPSVDGVVCRSADGKEVRYPVILTEIEKCIAFKLVRAFQQ 119
DVK YT+G F HAE RKSP VDG+V R+ GKEVRY L++ EK IA K+ +AF Q
Sbjct 410 FEDVKAYTIGENFCHAETRKSPVVDGIVRRNTHGKEVRYITELSDEEKTIAGKVSKAFSQ 469
Query 120 V 120
+
Sbjct 470 M 470
> Hs18547235
Length=440
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 1 EKPQEADNHDNWIYYPKNAGGGCKKLFRK 29
EKP A++H+ +IYYP +AG G ++LFRK
Sbjct 183 EKPVSAEDHNVYIYYPTSAGSGSQRLFRK 211
> AtMi069
Length=107
Score = 30.4 bits (67), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 15 YPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRK 46
YP+N G RK+QN +S IH +R+
Sbjct 11 YPQNVAGNMLSTLRKEQNQLTSVLTGIHILRR 42
> Hs22058025
Length=689
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 22/44 (50%), Gaps = 3/44 (6%)
Query 50 YLYEEFFPTFGTDVKVY---TVGPLFAHAEARKSPSVDGVVCRS 90
Y +E FP TD K + T GPL H EA +P V G R+
Sbjct 204 YNEKENFPANRTDYKAFIIKTAGPLPRHKEACAAPEVSGSQNRT 247
> At1g21340
Length=260
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 7/65 (10%)
Query 20 GGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTF--GTDVKVYTVGPLFAHAEA 77
GGGC+K R +QNS + ++ R DG+ ++ F + G+D+ + V FA
Sbjct 84 GGGCRKRSRSRQNSHKRF--GRNENRPDGLINQDDGFQSSPPGSDIDLAAV---FAQYVT 138
Query 78 RKSPS 82
+SPS
Sbjct 139 DRSPS 143
> 7303897
Length=372
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 25/57 (43%), Gaps = 3/57 (5%)
Query 38 FPD--IHQVRKDGVYLYEEFFPTFGTDVKVYTVGPLFAHAEARKSPSV-DGVVCRSA 91
PD I +K G+Y + TD+ Y++G L H A P + DG C A
Sbjct 175 LPDWMIECYKKIGIYAFGAVLSQLTTDIAKYSIGRLRPHFIAVCQPQMADGSTCDDA 231
> SPBC1685.05
Length=997
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 26/64 (40%), Gaps = 9/64 (14%)
Query 3 PQEADNHDNWIYYPKNAGGGCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFGTD 62
P +D N I+ PK FR + +S P V +G+Y E FP TD
Sbjct 3 PVSSDYRPNHIFLPK---------FRPDKITSECNTPQQQNVIAEGLYNVSEPFPIRNTD 53
Query 63 VKVY 66
+ Y
Sbjct 54 EERY 57
> 7294500
Length=440
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 11/41 (26%), Positives = 24/41 (58%), Gaps = 3/41 (7%)
Query 6 ADNHDNWIYYPK---NAGGGCKKLFRKQQNSSSSYFPDIHQ 43
+++ DN YY + +G GC ++F++ + S +F ++H
Sbjct 183 SEHADNEYYYAEKMGQSGAGCDRVFKECRRSLLQHFSELHH 223
> At1g47720
Length=272
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 34 SSSYFPDIHQVRKDGVYLYEEFF 56
SSS DI + +KD +YL++ FF
Sbjct 182 SSSLLDDIEESKKDEIYLWQVFF 204
> CE02574
Length=320
Score = 27.3 bits (59), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query 22 GCKKLFRKQQNSSSSYFPDIHQVRKDGVYLYEEFFPTFG 60
++ K +N S +F DI KDG ++EEF P F
Sbjct 94 NMEEHLEKSKNDSEIFF-DIVDTNKDGSIVWEEFEPHFS 131
Lambda K H
0.320 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40