bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6371_orf1
Length=119
Score E
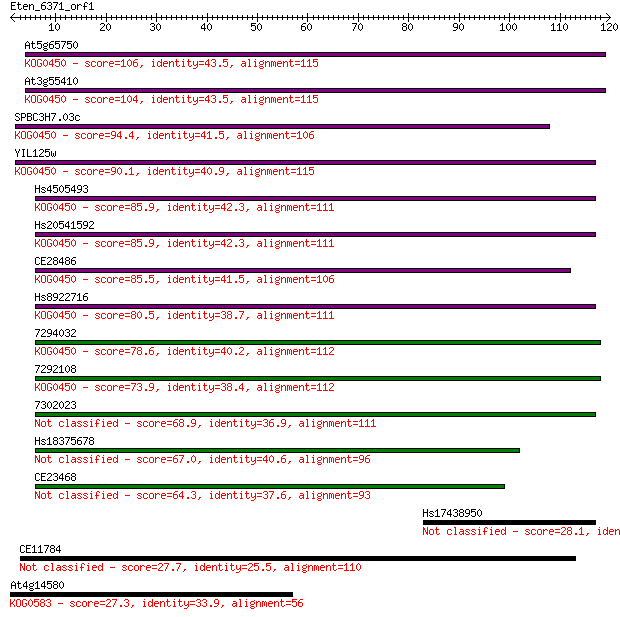
Sequences producing significant alignments: (Bits) Value
At5g65750 106 1e-23
At3g55410 104 4e-23
SPBC3H7.03c 94.4 5e-20
YIL125w 90.1 1e-18
Hs4505493 85.9 2e-17
Hs20541592 85.9 2e-17
CE28486 85.5 2e-17
Hs8922716 80.5 8e-16
7294032 78.6 3e-15
7292108 73.9 7e-14
7302023 68.9 2e-12
Hs18375678 67.0 8e-12
CE23468 64.3 5e-11
Hs17438950 28.1 3.7
CE11784 27.7 5.9
At4g14580 27.3 6.9
> At5g65750
Length=1025
Score = 106 bits (264), Expect = 1e-23, Method: Composition-based stats.
Identities = 50/115 (43%), Positives = 71/115 (61%), Gaps = 3/115 (2%)
Query 4 RVTGVRLSALRELGRRIFTIPEGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAY 63
R TGV+ L+ +G+ I T PE F PH + ++ +QR +E E +D+G E LA+
Sbjct 593 RNTGVKPEILKNVGKAISTFPENFKPHRGVKRVYEQRAQMIESGEG---IDWGLGEALAF 649
Query 64 ASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCIFDSLKDLNLPHTITI 118
A+L+ +G H+RL+GQD +RGTFSHRH+VLHDQ +YC D L P T+
Sbjct 650 ATLVVEGNHVRLSGQDVERGTFSHRHSVLHDQETGEEYCPLDHLIKNQDPEMFTV 704
> At3g55410
Length=1009
Score = 104 bits (259), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 50/115 (43%), Positives = 72/115 (62%), Gaps = 3/115 (2%)
Query 4 RVTGVRLSALRELGRRIFTIPEGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAY 63
R TGV+ L+ +G+ I ++PE F PH + K+ +QR +E E +D+ AE LA+
Sbjct 589 RNTGVKPEILKTVGKAISSLPENFKPHRAVKKVYEQRAQMIESGEG---VDWALAEALAF 645
Query 64 ASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCIFDSLKDLNLPHTITI 118
A+L+ +G H+RL+GQD +RGTFSHRH+VLHDQ +YC D L P T+
Sbjct 646 ATLVVEGNHVRLSGQDVERGTFSHRHSVLHDQETGEEYCPLDHLIMNQDPEMFTV 700
> SPBC3H7.03c
Length=1009
Score = 94.4 bits (233), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 66/106 (62%), Gaps = 3/106 (2%)
Query 2 PPRVTGVRLSALRELGRRIFTIPEGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENL 61
P TGV + L+++G+ ++T+PEGF H + +I R ++ E +D AE L
Sbjct 596 PSYPTGVNIDTLKQIGKALYTLPEGFDAHRNLKRILNNRNKSISSGEG---IDMPTAEAL 652
Query 62 AYASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCIFDSL 107
A+ +LL +G H+R++GQD +RGTFS RHAVLHDQ+ E Y + L
Sbjct 653 AFGTLLEEGHHVRVSGQDVERGTFSQRHAVLHDQSSENVYIPLNHL 698
> YIL125w
Length=1014
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/115 (40%), Positives = 68/115 (59%), Gaps = 3/115 (2%)
Query 2 PPRVTGVRLSALRELGRRIFTIPEGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENL 61
P T V S L+ELG+ + + PEGF H + +I K R ++E E +D+ E L
Sbjct 598 PHEPTNVPESTLKELGKVLSSWPEGFEVHKNLKRILKNRGKSIETGEG---IDWATGEAL 654
Query 62 AYASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCIFDSLKDLNLPHTI 116
A+ +L+ DG ++R++G+D +RGTFS RHAVLHDQ EA Y +L + TI
Sbjct 655 AFGTLVLDGQNVRVSGEDVERGTFSQRHAVLHDQQSEAIYTPLSTLNNEKADFTI 709
> Hs4505493
Length=1002
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 47/113 (41%), Positives = 68/113 (60%), Gaps = 6/113 (5%)
Query 6 TGVRLSALRELGRRIFTIP-EGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAYA 64
TG+ L +G ++P E F H +++I K R V+ +T+D+ AE +A+
Sbjct 607 TGLTEDILTHIGNVASSVPVENFTIHGGLSRILKTRGEMVKN----RTVDWALAEYMAFG 662
Query 65 SLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCI-FDSLKDLNLPHTI 116
SLL +G HIRL+GQD +RGTFSHRH VLHDQ V+ + CI + L P+T+
Sbjct 663 SLLKEGIHIRLSGQDVERGTFSHRHHVLHDQNVDKRTCIPMNHLWPNQAPYTV 715
> Hs20541592
Length=1023
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 47/113 (41%), Positives = 68/113 (60%), Gaps = 6/113 (5%)
Query 6 TGVRLSALRELGRRIFTIP-EGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAYA 64
TG+ L +G ++P E F H +++I K R V+ +T+D+ AE +A+
Sbjct 607 TGLTEDILTHIGNVASSVPVENFTIHGGLSRILKTRGEMVKN----RTVDWALAEYMAFG 662
Query 65 SLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCI-FDSLKDLNLPHTI 116
SLL +G HIRL+GQD +RGTFSHRH VLHDQ V+ + CI + L P+T+
Sbjct 663 SLLKEGIHIRLSGQDVERGTFSHRHHVLHDQNVDKRTCIPMNHLWPNQAPYTV 715
> CE28486
Length=1029
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 64/106 (60%), Gaps = 6/106 (5%)
Query 6 TGVRLSALRELGRRIFTIPEGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAYAS 65
TG+ + ++ + PEGF H + + K R ++ N +LD+ E LA+ S
Sbjct 608 TGIEQENIEQIIGKFSQYPEGFNLHRGLERTLKGRQQMLKDN----SLDWACGEALAFGS 663
Query 66 LLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCIFDSLKDLN 111
LL +G H+RL+GQD QRGTFSHRH VLHDQ V+ + I++ L DL+
Sbjct 664 LLKEGIHVRLSGQDVQRGTFSHRHHVLHDQKVDQK--IYNPLNDLS 707
> Hs8922716
Length=1010
Score = 80.5 bits (197), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 43/113 (38%), Positives = 67/113 (59%), Gaps = 6/113 (5%)
Query 6 TGVRLSALRELGRRIFTIP-EGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAYA 64
TG+ L +G ++P E F H +++I + R + +T+D+ AE +A+
Sbjct 594 TGIPEDMLTHIGSVASSVPLEDFKIHTGLSRILRGRADMTKN----RTVDWALAEYMAFG 649
Query 65 SLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCI-FDSLKDLNLPHTI 116
SLL +G H+RL+GQD +RGTFSHRH VLHDQ V+ + C+ + L P+T+
Sbjct 650 SLLKEGIHVRLSGQDVERGTFSHRHHVLHDQEVDRRTCVPMNHLWPDQAPYTV 702
> 7294032
Length=1001
Score = 78.6 bits (192), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 64/116 (55%), Gaps = 8/116 (6%)
Query 6 TGVRLSALRELGRRIFTIPEG---FVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLA 62
TGV+ L +G R + P FV H + ++ R A V DEK D+ E +A
Sbjct 598 TGVKEETLIHIGNRFSSPPPNAAEFVIHKGLLRVLAARKAMV----DEKVADWALGEAMA 653
Query 63 YASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVE-AQYCIFDSLKDLNLPHTIT 117
+ SLL +G H+RL+GQD +RGTFSHRH VLH Q V+ A Y + P++++
Sbjct 654 FGSLLKEGIHVRLSGQDVERGTFSHRHHVLHHQLVDKATYNSLQHMYPDQAPYSVS 709
> 7292108
Length=1075
Score = 73.9 bits (180), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 43/116 (37%), Positives = 61/116 (52%), Gaps = 8/116 (6%)
Query 6 TGVRLSALRELGRRIFTIP---EGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLA 62
TG+ L+ +G T P F H I +I QR V+ +K D+ E A
Sbjct 621 TGISTDTLKTIGNMFSTPPPPEHKFETHKGILRILAQRTQMVQ----DKVADWSLGEAFA 676
Query 63 YASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVE-AQYCIFDSLKDLNLPHTIT 117
+ SLL +G H+RL+GQD +RGTFSHRH VLH Q+ + Y D L P++++
Sbjct 677 FGSLLKEGIHVRLSGQDVERGTFSHRHHVLHHQSEDKVVYNSLDHLYPDQAPYSVS 732
> 7302023
Length=901
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 41/116 (35%), Positives = 64/116 (55%), Gaps = 7/116 (6%)
Query 6 TGVRLSALRELGRRIFTIPEGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAYAS 65
TG+ S L +G++ T PE F H + +K A ++ E+ +D+ AE LA S
Sbjct 507 TGLDYSLLHYIGQQSVTFPEDFNIHPHL--LKTHVNARLKKLENGVKIDWSTAEALAIGS 564
Query 66 LLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCIFDSLK-----DLNLPHTI 116
L+ G ++R++G+D RGTFSHRHA+L DQ + +S++ L L H+I
Sbjct 565 LMYQGHNVRISGEDVGRGTFSHRHAMLVDQQTNEMFIPLNSMEGGNGGKLELAHSI 620
> Hs18375678
Length=919
Score = 67.0 bits (162), Expect = 8e-12, Method: Composition-based stats.
Identities = 39/96 (40%), Positives = 54/96 (56%), Gaps = 2/96 (2%)
Query 6 TGVRLSALRELGRRIFTIPEGFVPHATIAKIKKQRLAAVEGNEDEKTLDFGAAENLAYAS 65
TGV L LR +G + +P H+ + K Q + +E D LD+ AE LA S
Sbjct 524 TGVPLDLLRFVGMKSVEVPRELQMHSHLLKTHVQ--SRMEKMMDGIKLDWATAEALALGS 581
Query 66 LLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQY 101
LL+ GF++RL+GQD RGTFS RHA++ Q + Y
Sbjct 582 LLAQGFNVRLSGQDVGRGTFSQRHAMVVCQETDDTY 617
> CE23468
Length=906
Score = 64.3 bits (155), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 55/97 (56%), Gaps = 10/97 (10%)
Query 6 TGVRLSALRELGRRIFTIPEGFVPHATIAKI----KKQRLAAVEGNEDEKTLDFGAAENL 61
TGV LR +G +PE F H + K+ + Q++ EG +D+ AE +
Sbjct 509 TGVATDLLRFIGAGSVKVPEDFDTHKHLYKMHIDSRMQKMQTGEG------IDWATAEAM 562
Query 62 AYASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVE 98
A+ S+L +G +R++GQD RGTF HRHA++ DQ+ +
Sbjct 563 AFGSILLEGNDVRISGQDVGRGTFCHRHAMMVDQSTD 599
> Hs17438950
Length=1000
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 2/36 (5%)
Query 83 GTFSHRHAVLHDQAVE--AQYCIFDSLKDLNLPHTI 116
GTF HA L DQ +E +QY LK+L+L H +
Sbjct 296 GTFIFCHAYLADQDMECLSQYPSLSQLKELHLIHIL 331
> CE11784
Length=338
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 51/115 (44%), Gaps = 6/115 (5%)
Query 3 PRVTGVRLSALRELGRRIFTIPEG--FVPHATIAK---IKKQRLAAVEGNEDEKTLDFGA 57
P+ R E+G R F+ P+ F + ++ + I LA +G L+F +
Sbjct 53 PKCQFCRFKKCLEIGMR-FSEPKQQLFNLNNSVDQDLIILLGNLAVKDGVHYSNFLNFYS 111
Query 58 AENLAYASLLSDGFHIRLAGQDAQRGTFSHRHAVLHDQAVEAQYCIFDSLKDLNL 112
E+ + +L+D ++L + + T SH L AQ+ FD +K ++L
Sbjct 112 LEDPSMDDILADRSRMKLMKRTLEIKTKSHEWTFLGSYYKIAQFLDFDFVKTMSL 166
> At4g14580
Length=426
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 4/57 (7%)
Query 1 SPPRVTGVRLSALRELGRRIFTIPEGFVPHATIAK-IKKQRLAAVEGNEDEKTLDFG 56
SP ++TG L ELGRR+ + G +A+ I L A++ + +KT+D G
Sbjct 8 SPEKITGTVLLGKYELGRRLGS---GSFAKVHVARSISTGELVAIKIIDKQKTIDSG 61
Lambda K H
0.322 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40