bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6379_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
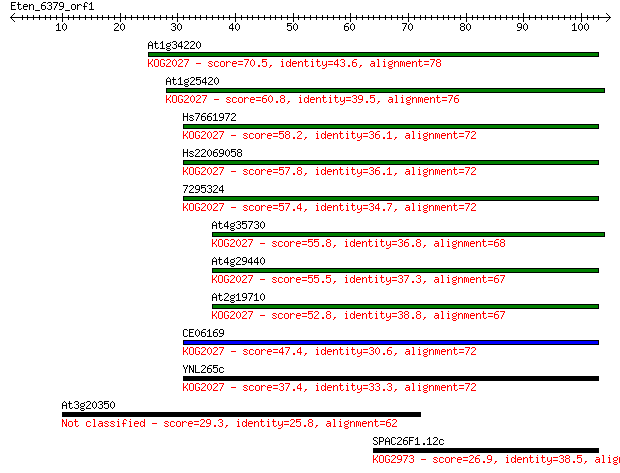
At1g34220 70.5 7e-13
At1g25420 60.8 6e-10
Hs7661972 58.2 4e-09
Hs22069058 57.8 5e-09
7295324 57.4 7e-09
At4g35730 55.8 2e-08
At4g29440 55.5 2e-08
At2g19710 52.8 2e-07
CE06169 47.4 6e-06
YNL265c 37.4 0.006
At3g20350 29.3 1.7
SPAC26F1.12c 26.9 9.5
> At1g34220
Length=619
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 47/78 (60%), Gaps = 0/78 (0%)
Query 25 FDKAVCKASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRL 84
F A CK LK+ I R ++ NR + I+ RREIA L G+E AR++ EH++RE ++
Sbjct 12 FKAAKCKTLLKLTIPRIKLIRNRREAQIKQMRREIAKLLETGQEATARIRVEHIIREEKM 71
Query 85 ERAMEILVTVCELLISRL 102
A EIL CEL+ RL
Sbjct 72 MAAQEILELFCELIAVRL 89
> At1g25420
Length=323
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 49/76 (64%), Gaps = 0/76 (0%)
Query 28 AVCKASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERA 87
A CK SL +AI+R ++ N+ ++ ++EIA FL+ G+E AR++ EH++RE L A
Sbjct 15 AKCKTSLNLAIARMKLLQNKRDMQLKHMKKEIAHFLQAGQEPIARIRVEHVIREMNLWAA 74
Query 88 MEILVTVCELLISRLS 103
EIL CE +++R+
Sbjct 75 YEILELFCEFILARVP 90
> Hs7661972
Length=364
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 31 KASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEI 90
+ +L++ I+R ++ + + R+EIA +L G++E AR++ EH++RE L AMEI
Sbjct 12 RVNLRLVINRLKLLEKKKTELAQKARKEIADYLAAGKDERARIRVEHIIREDYLVEAMEI 71
Query 91 LVTVCELLISRL 102
L C+LL++R
Sbjct 72 LELYCDLLLARF 83
> Hs22069058
Length=360
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 31 KASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEI 90
+ +L++ I+R ++ + + R+EIA +L G++E AR++ EH++RE L AMEI
Sbjct 12 RVNLRLVINRLKLLEKKKTELAQKARKEIADYLAAGKDERARIRVEHIIREDYLVEAMEI 71
Query 91 LVTVCELLISRL 102
L C+LL++R
Sbjct 72 LELYCDLLLARF 83
> 7295324
Length=417
Score = 57.4 bits (137), Expect = 7e-09, Method: Composition-based stats.
Identities = 25/72 (34%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 31 KASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEI 90
K +L++A++R ++ + + R+EIA +L G+ E AR++ EH++RE L AME+
Sbjct 12 KTNLRLALNRLKLLEKKKAELTQKSRKEIADYLATGKTERARIRVEHIIREDYLVEAMEM 71
Query 91 LVTVCELLISRL 102
+ C+LL++R
Sbjct 72 VEMYCDLLLARF 83
> At4g35730
Length=430
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 46/68 (67%), Gaps = 0/68 (0%)
Query 36 MAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEILVTVC 95
MA++R ++ N+ ++ RR+IA L+ G++ AR++ EH++RE ++ A EI+ C
Sbjct 1 MAVARIKLIRNKRLVVVKQMRRDIAVLLQSGQDATARIRVEHVIREQNIQAANEIIELFC 60
Query 96 ELLISRLS 103
EL++SRL+
Sbjct 61 ELIVSRLT 68
> At4g29440
Length=1071
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 42/67 (62%), Gaps = 0/67 (0%)
Query 36 MAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEILVTVC 95
MA SR ++ N+ I+ RRE+A L G+ + A+++ EH++RE + A E++ C
Sbjct 1 MAASRLKILKNKKDTQIKQLRRELAHLLESGQTQTAKIRVEHVVREEKTVAAYELVGIYC 60
Query 96 ELLISRL 102
ELL++RL
Sbjct 61 ELLVARL 67
> At2g19710
Length=918
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 41/67 (61%), Gaps = 0/67 (0%)
Query 36 MAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEILVTVC 95
MA SR ++ N+ + I+ RRE+A L G+ AR++ EH++RE + A E++ C
Sbjct 1 MANSRLKILKNKKEIQIKQLRRELAQLLESGQTPTARIRVEHVVREEKTVAAYELIGIYC 60
Query 96 ELLISRL 102
ELL+ RL
Sbjct 61 ELLVVRL 67
> CE06169
Length=432
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 41/72 (56%), Gaps = 0/72 (0%)
Query 31 KASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEI 90
K +L++ I+R ++ + R EIA ++ + + AR++ EH++RE + A EI
Sbjct 13 KTNLRLGINRLQLLGKKKTEMAMKARTEIADYIAANKPDRARIRVEHIIREDYVVEAFEI 72
Query 91 LVTVCELLISRL 102
L C+LL++R
Sbjct 73 LEMYCDLLLARF 84
> YNL265c
Length=298
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 0/72 (0%)
Query 31 KASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREEGARLKAEHLLREYRLERAMEI 90
K LKM I R R + Q + RR++A L +E+ A + E L+ + +EI
Sbjct 13 KTCLKMCIQRLRYAQEKQQAIAKQSRRQVAQLLLTNKEQKAHYRVETLIHDDIHIELLEI 72
Query 91 LVTVCELLISRL 102
L CELL++R+
Sbjct 73 LELYCELLLARV 84
> At3g20350
Length=673
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 10 RRLATNMKMRWPFERFDKAVCKASLKMAISRARMCTNRLQNSIRLQRREIAGFLREGREE 69
++ TN+K W ++ + +S+++ + AR C L++ R Q++++ FL++ EE
Sbjct 197 HQIYTNVK--WNNQQVNDVSLASSIELKLQEARACIKDLESEKRSQKKKLEQFLKKVSEE 254
Query 70 GA 71
A
Sbjct 255 RA 256
> SPAC26F1.12c
Length=356
Score = 26.9 bits (58), Expect = 9.5, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 64 REGREEGARLKAEHLLREYRLERAMEILVTVCELLISRL 102
REGRE R K ++RE L E + VC+ L+ L
Sbjct 291 REGREHMRRRKVYPIIRELHLNVDDEEIREVCDQLVQML 329
Lambda K H
0.330 0.137 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40